Gene Page: KMT2A
Summary ?
| GeneID | 4297 |
| Symbol | KMT2A |
| Synonyms | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL-AF9|MLL/GAS7|MLL1|MLL1A|TET1-MLL|TRX1|WDSTS |
| Description | lysine methyltransferase 2A |
| Reference | MIM:159555|HGNC:HGNC:7132|Ensembl:ENSG00000118058|HPRD:01162|Vega:OTTHUMG00000166337 |
| Gene type | protein-coding |
| Map location | 11q23 |
| Pascal p-value | 6.823E-4 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
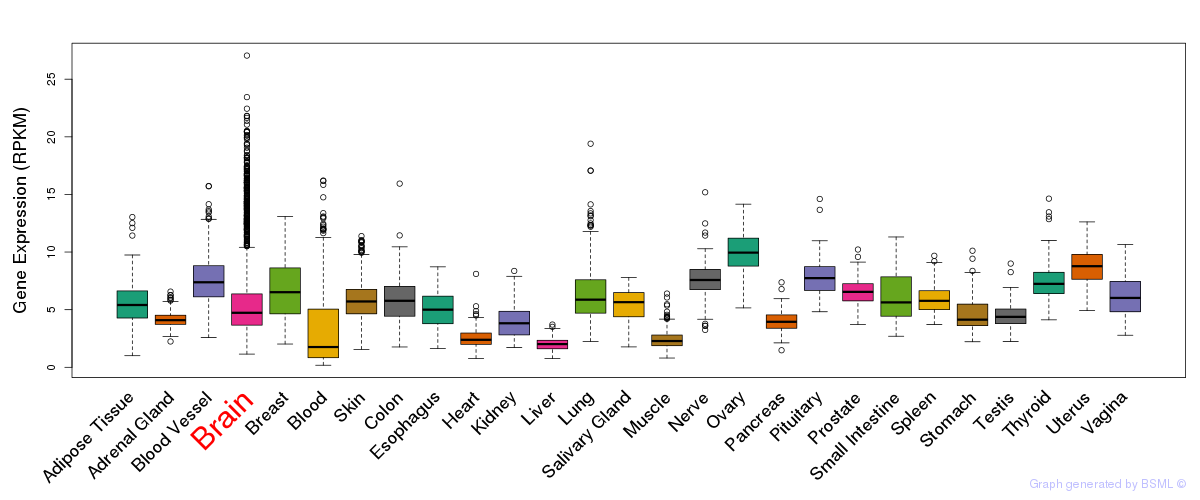
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ERBB2 | 0.88 | 0.83 |
| MRC2 | 0.88 | 0.84 |
| EPHB4 | 0.87 | 0.85 |
| FSTL1 | 0.86 | 0.79 |
| PDPN | 0.85 | 0.68 |
| CMTM3 | 0.84 | 0.72 |
| IQGAP2 | 0.83 | 0.69 |
| CPXM1 | 0.83 | 0.67 |
| MSN | 0.82 | 0.76 |
| SMO | 0.82 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.41 | -0.49 |
| CA11 | -0.40 | -0.49 |
| CA4 | -0.40 | -0.43 |
| CACNA1F | -0.37 | -0.43 |
| CCNI2 | -0.37 | -0.40 |
| SPDEF | -0.36 | -0.44 |
| AF347015.27 | -0.36 | -0.44 |
| EPHX4 | -0.36 | -0.46 |
| ASPHD1 | -0.36 | -0.32 |
| TUBA1 | -0.36 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003700 | transcription factor activity | NAS | 10821850 | |
| GO:0003702 | RNA polymerase II transcription factor activity | TAS | 10821850 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008168 | methyltransferase activity | IEA | - | |
| GO:0018024 | histone-lysine N-methyltransferase activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | IDA | 11313484 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006461 | protein complex assembly | IDA | 15199122 | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 10821850 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0035162 | embryonic hemopoiesis | TAS | 15618964 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 15640349 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASH2L | ASH2 | ASH2L1 | ASH2L2 | Bre2 | ash2 (absent, small, or homeotic)-like (Drosophila) | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| AVP | ADH | ARVP | AVP-NPII | AVRP | VP | arginine vasopressin | Biochemical Activity | BioGRID | 14607844 |
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | - | HPRD,BioGRID | 12829790 |
| CBX4 | NBP16 | PC2 | hPC2 | chromobox homolog 4 (Pc class homolog, Drosophila) | Reconstituted Complex | BioGRID | 12829790 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 11259575 |12205094 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12829790 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD | 12023017 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| HCFC2 | FLJ94012 | HCF-2 | HCF2 | host cell factor C2 | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12829790 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | MLL1 interacts with HOXA9 promoter. | BIND | 15960974 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | MLL1 interacts with HOXA9. | BIND | 15960974 |
| HOXC8 | HOX3 | HOX3A | homeobox C8 | MLL1 interacts with HOXC8 promoter. | BIND | 15960974 |
| HOXC8 | HOX3 | HOX3A | homeobox C8 | MLL1 interacts with HOXC8. | BIND | 15960974 |
| INS | ILPR | IRDN | insulin | Biochemical Activity | BioGRID | 14607844 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | Affinity Capture-Western | BioGRID | 14607843 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | - | HPRD,BioGRID | 11313484 |12393701 |12482972 |
| MYST1 | FLJ14040 | KAT8 | MOF | hMOF | MYST histone acetyltransferase 1 | MOF interacts with MLL. | BIND | 15960975 |
| OXT | MGC126890 | MGC126892 | OT | OT-NPI | oxytocin, prepropeptide | Biochemical Activity | BioGRID | 14607844 |
| PLXNB1 | KIAA0407 | MGC149167 | PLEXIN-B1 | PLXN5 | SEP | plexin B1 | - | HPRD | 12123608 |
| PPIE | CYP-33 | MGC111222 | MGC3736 | peptidylprolyl isomerase E (cyclophilin E) | - | HPRD,BioGRID | 11313484 |
| PPP1R15A | GADD34 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | - | HPRD,BioGRID | 10490642 |
| RBBP5 | RBQ3 | SWD1 | retinoblastoma binding protein 5 | RbBP5 interacts with MLL. | BIND | 15960975 |
| RBBP5 | RBQ3 | SWD1 | retinoblastoma binding protein 5 | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| RNF2 | BAP-1 | BAP1 | DING | HIPI3 | RING1B | RING2 | ring finger protein 2 | Ring2 interacts with MLL. | BIND | 15960975 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-Western | BioGRID | 10490642 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | - | HPRD | 10821850 |
| WDR5 | BIG-3 | SWD3 | WD repeat domain 5 | WDR5 interacts with MLL. This interaction was modeled on a demonstrated interaction between WDR5 from an unspecified species and human MLL. | BIND | 15960975 |
| WDR5 | BIG-3 | SWD3 | WD repeat domain 5 | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 2689 | 2695 | m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 2219 | 2225 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 2691 | 2697 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 2486 | 2492 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-136 | 2360 | 2366 | 1A | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-137 | 2167 | 2174 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-142-3p | 1242 | 1248 | m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-148/152 | 2583 | 2590 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| hsa-miR-148a | UCAGUGCACUACAGAACUUUGU | ||||
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-150 | 537 | 544 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-182 | 2857 | 2864 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-193 | 2624 | 2630 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-218 | 2031 | 2037 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-221/222 | 954 | 961 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-27 | 2691 | 2697 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 1364 | 1371 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-338 | 2279 | 2285 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-342 | 2493 | 2499 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-369-3p | 1478 | 1484 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-370 | 2372 | 2378 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-377 | 2492 | 2498 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-380-5p | 2808 | 2815 | 1A,m8 | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-381 | 1408 | 1414 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-494 | 1290 | 1296 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-495 | 2563 | 2569 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 1395 | 1401 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1445 | 1452 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG | ||||
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 2858 | 2864 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.