Gene Page: NTM
Summary ?
| GeneID | 50863 |
| Symbol | NTM |
| Synonyms | HNT|IGLON2|NTRI |
| Description | neurotrimin |
| Reference | MIM:607938|HGNC:HGNC:17941|Ensembl:ENSG00000182667|HPRD:06398|Vega:OTTHUMG00000066364 |
| Gene type | protein-coding |
| Map location | 11q25 |
| Pascal p-value | 0.008 |
| Fetal beta | -1.22 |
| DMG | 1 (# studies) |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:Ripke_2013 | Genome-wide Association Study | Multi-stage GWAS, Sweden population and PGC2. 24 leading SNPs | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22887491 | 11 | 131618277 | NTM | 2.43E-5 | 0.005 | 0.067 | DMG:Montano_2016 |
Section II. Transcriptome annotation
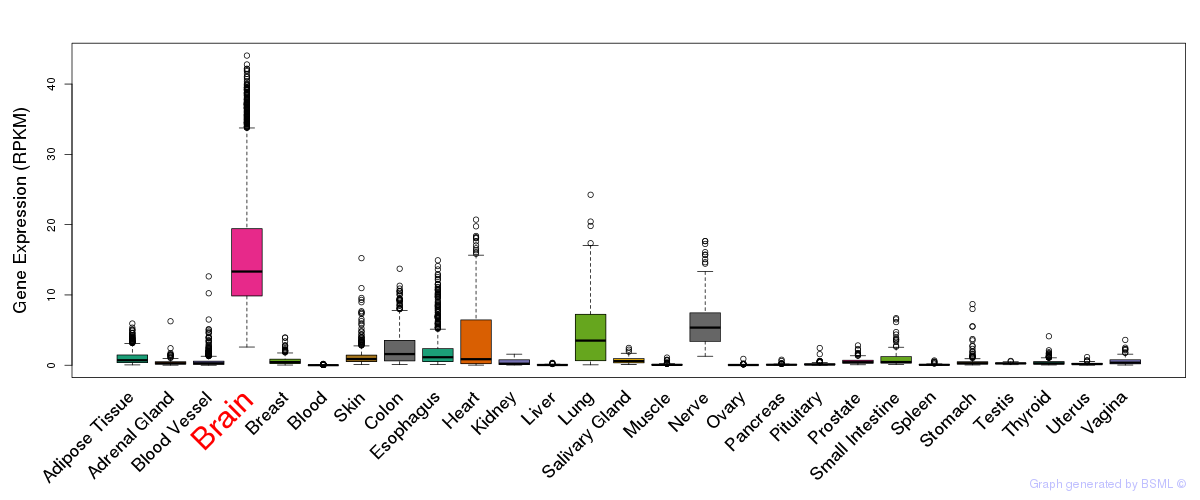
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008038 | neuron recognition | TAS | neuron (GO term level: 4) | 7891157 |
| GO:0007155 | cell adhesion | NAS | 7891157 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| YAGUE PRETUMOR DRUG RESISTANCE DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3K27ME3 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR DN | 36 | 19 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 882 | 888 | m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-151 | 678 | 685 | 1A,m8 | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-188 | 629 | 635 | m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-28 | 1177 | 1183 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-30-5p | 789 | 795 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-346 | 921 | 927 | 1A | hsa-miR-346brain | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-361 | 669 | 675 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-369-3p | 1017 | 1023 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1017 | 1023 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-485-3p | 1489 | 1496 | 1A,m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-495 | 1048 | 1054 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-504 | 1080 | 1086 | m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
| miR-505 | 1129 | 1135 | m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.