Gene Page: POLR3F
Summary ?
| GeneID | 10621 |
| Symbol | POLR3F |
| Synonyms | RPC39|RPC6 |
| Description | polymerase (RNA) III subunit F |
| Reference | HGNC:HGNC:15763|Ensembl:ENSG00000132664|HPRD:15156|Vega:OTTHUMG00000031971 |
| Gene type | protein-coding |
| Map location | 20p11.23 |
| Pascal p-value | 0.684 |
| Sherlock p-value | 0.831 |
| Fetal beta | 0.1 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.046 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14535006 | 20 | 18448030 | POLR3F | 7.14E-9 | -0.025 | 3.59E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6075348 | chr20 | 18473041 | POLR3F | 10621 | 0.06 | cis | ||
| rs2247746 | chr20 | 18477613 | POLR3F | 10621 | 0.2 | cis | ||
| rs7262824 | chr20 | 18483104 | POLR3F | 10621 | 0.09 | cis | ||
| rs1022681 | chr20 | 18487013 | POLR3F | 10621 | 0.06 | cis | ||
| rs1022684 | chr20 | 18487505 | POLR3F | 10621 | 0.15 | cis | ||
| rs1109030 | chr20 | 18506676 | POLR3F | 10621 | 0.13 | cis | ||
| rs1555354 | chr20 | 18506824 | POLR3F | 10621 | 0.04 | cis | ||
| rs911011 | chr20 | 18717017 | POLR3F | 10621 | 0.14 | cis |
Section II. Transcriptome annotation
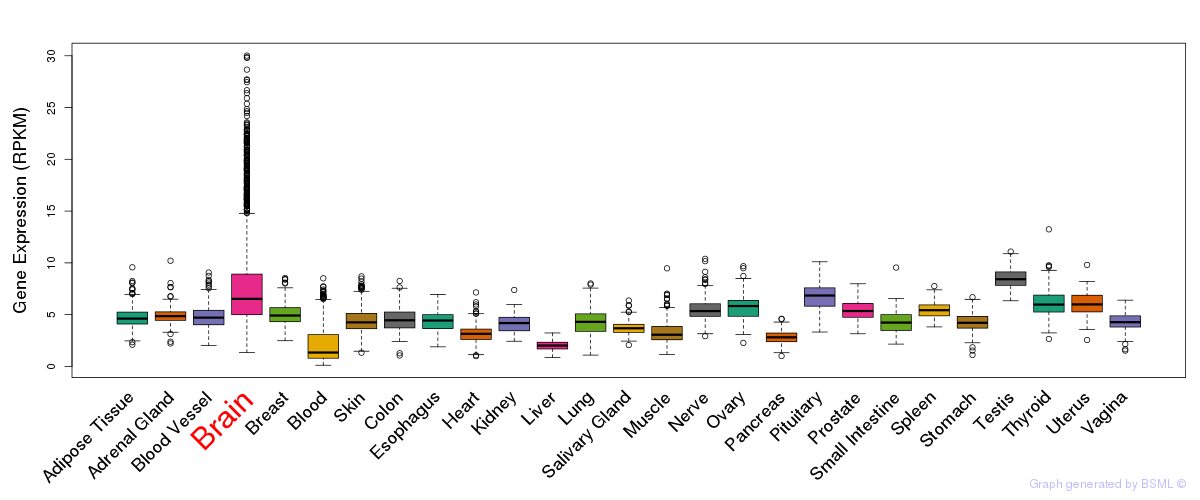
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003899 | DNA-directed RNA polymerase activity | NAS | 9171375 | |
| GO:0005515 | protein binding | IPI | 16169070 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006359 | regulation of transcription from RNA polymerase III promoter | NAS | 9171375 | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005666 | DNA-directed RNA polymerase III complex | NAS | 10623476 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BDP1 | DKFZp686C01233 | DKFZp686K0831 | HSA238520 | KIAA1241 | KIAA1689 | TAF3B1 | TFC5 | TFIIIB'' | TFIIIB150 | TFIIIB90 | TFNR | B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB | - | HPRD | 9171375 |
| BRF1 | BRF | FLJ42674 | FLJ43034 | GTF3B | MGC105048 | TAF3B2 | TAF3C | TAFIII90 | TF3B90 | TFIIIB90 | hBRF | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae) | - | HPRD | 9171375 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| GTF3C4 | FLJ21002 | KAT12 | MGC138450 | TFIII90 | TFIIIC90 | TFIIICdelta | TFiiiC2-90 | general transcription factor IIIC, polypeptide 4, 90kDa | - | HPRD,BioGRID | 10523658 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | Two-hybrid | BioGRID | 16169070 |
| POLR3C | RPC3 | RPC62 | polymerase (RNA) III (DNA directed) polypeptide C (62kD) | - | HPRD,BioGRID | 9171375 |
| POLR3D | BN51T | RPC4 | RPC53 | TSBN51 | polymerase (RNA) III (DNA directed) polypeptide D, 44kDa | Affinity Capture-MS | BioGRID | 12391170 |
| POLR3G | RPC32 | RPC7 | polymerase (RNA) III (DNA directed) polypeptide G (32kD) | - | HPRD | 9171375 |
| PRPF4B | KIAA0536 | PR4H | PRP4 | PRP4H | PRP4K | dJ1013A10.1 | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | Two-hybrid | BioGRID | 16169070 |
| STC2 | STC-2 | STCRP | stanniocalcin 2 | Two-hybrid | BioGRID | 16169070 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 9171375 |
| WDR89 | C14orf150 | MGC9907 | MSTP050 | WD repeat domain 89 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG RNA POLYMERASE | 29 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | 23 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION TERMINATION | 19 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III CHAIN ELONGATION | 17 | 8 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 122 | 129 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-503 | 123 | 129 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.