Gene Page: GAS2L1
Summary ?
| GeneID | 10634 |
| Symbol | GAS2L1 |
| Synonyms | GAR22 |
| Description | growth arrest specific 2 like 1 |
| Reference | MIM:602128|HGNC:HGNC:16955|Ensembl:ENSG00000185340|HPRD:03676|Vega:OTTHUMG00000151108 |
| Gene type | protein-coding |
| Map location | 22q12.2 |
| Pascal p-value | 0.204 |
| Sherlock p-value | 0.581 |
| Fetal beta | -1.219 |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs909798 | 22 | 29654004 | GAS2L1 | ENSG00000185340.11 | 2.729E-7 | 0 | -48568 | gtex_brain_putamen_basal |
| rs2857466 | 22 | 29657604 | GAS2L1 | ENSG00000185340.11 | 3.188E-7 | 0 | -44968 | gtex_brain_putamen_basal |
| rs2301291 | 22 | 29664423 | GAS2L1 | ENSG00000185340.11 | 5.214E-7 | 0 | -38149 | gtex_brain_putamen_basal |
| rs9613857 | 22 | 29679713 | GAS2L1 | ENSG00000185340.11 | 2.208E-7 | 0 | -22859 | gtex_brain_putamen_basal |
| rs3761426 | 22 | 29688632 | GAS2L1 | ENSG00000185340.11 | 2.227E-7 | 0 | -13940 | gtex_brain_putamen_basal |
| rs3747142 | 22 | 29693990 | GAS2L1 | ENSG00000185340.11 | 1.656E-7 | 0 | -8582 | gtex_brain_putamen_basal |
| rs2301586 | 22 | 29702910 | GAS2L1 | ENSG00000185340.11 | 4.668E-7 | 0 | 338 | gtex_brain_putamen_basal |
| rs9613859 | 22 | 29703409 | GAS2L1 | ENSG00000185340.11 | 2.774E-7 | 0 | 837 | gtex_brain_putamen_basal |
| rs142333573 | 22 | 29706244 | GAS2L1 | ENSG00000185340.11 | 2.501E-7 | 0 | 3672 | gtex_brain_putamen_basal |
| rs9613860 | 22 | 29707955 | GAS2L1 | ENSG00000185340.11 | 2.025E-7 | 0 | 5383 | gtex_brain_putamen_basal |
| rs3216803 | 22 | 29708796 | GAS2L1 | ENSG00000185340.11 | 3.082E-7 | 0 | 6224 | gtex_brain_putamen_basal |
| rs71700945 | 22 | 29712522 | GAS2L1 | ENSG00000185340.11 | 1.092E-7 | 0 | 9950 | gtex_brain_putamen_basal |
| rs6006093 | 22 | 29719167 | GAS2L1 | ENSG00000185340.11 | 2.194E-8 | 0 | 16595 | gtex_brain_putamen_basal |
| rs9613863 | 22 | 29721902 | GAS2L1 | ENSG00000185340.11 | 3.548E-8 | 0 | 19330 | gtex_brain_putamen_basal |
| rs9613864 | 22 | 29728854 | GAS2L1 | ENSG00000185340.11 | 4.309E-8 | 0 | 26282 | gtex_brain_putamen_basal |
| rs3747146 | 22 | 29735310 | GAS2L1 | ENSG00000185340.11 | 9.375E-9 | 0 | 32738 | gtex_brain_putamen_basal |
| rs715589 | 22 | 29743779 | GAS2L1 | ENSG00000185340.11 | 7.725E-9 | 0 | 41207 | gtex_brain_putamen_basal |
| rs737883 | 22 | 29746253 | GAS2L1 | ENSG00000185340.11 | 5.014E-6 | 0 | 43681 | gtex_brain_putamen_basal |
| rs174650 | 22 | 29883892 | GAS2L1 | ENSG00000185340.11 | 3.158E-6 | 0 | 181320 | gtex_brain_putamen_basal |
| rs165743 | 22 | 29884277 | GAS2L1 | ENSG00000185340.11 | 2.971E-6 | 0 | 181705 | gtex_brain_putamen_basal |
| rs5763269 | 22 | 29885473 | GAS2L1 | ENSG00000185340.11 | 3.214E-6 | 0 | 182901 | gtex_brain_putamen_basal |
| rs165923 | 22 | 29885861 | GAS2L1 | ENSG00000185340.11 | 3.228E-6 | 0 | 183289 | gtex_brain_putamen_basal |
| rs165625 | 22 | 29886413 | GAS2L1 | ENSG00000185340.11 | 3.242E-6 | 0 | 183841 | gtex_brain_putamen_basal |
| rs165894 | 22 | 29887623 | GAS2L1 | ENSG00000185340.11 | 2.278E-6 | 0 | 185051 | gtex_brain_putamen_basal |
| rs58197542 | 22 | 30121774 | GAS2L1 | ENSG00000185340.11 | 2.381E-6 | 0 | 419202 | gtex_brain_putamen_basal |
| rs36573 | 22 | 30203919 | GAS2L1 | ENSG00000185340.11 | 3.011E-6 | 0 | 501347 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
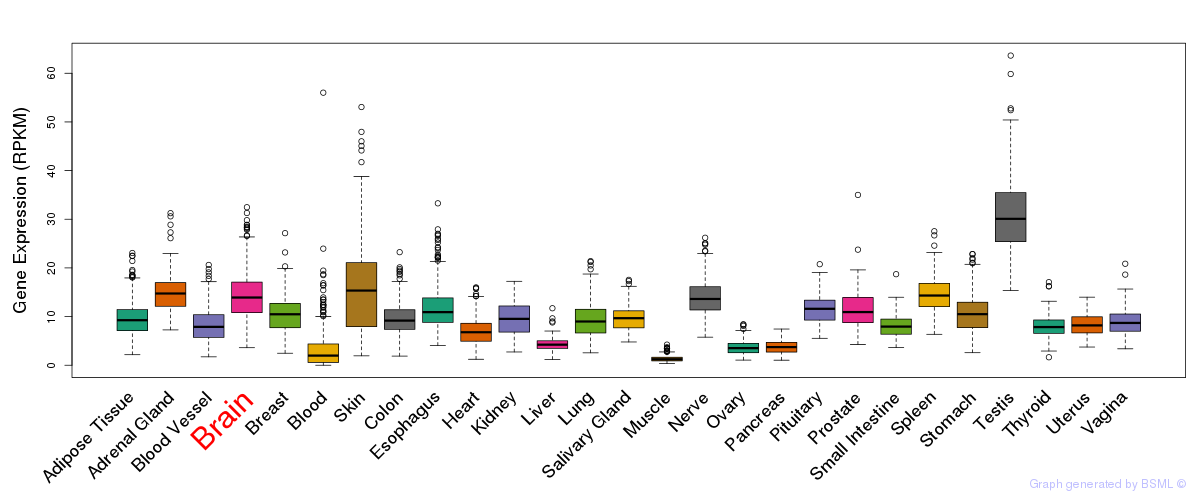
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007050 | cell cycle arrest | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS DN | 36 | 21 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 DN | 46 | 31 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID MN1 NETWORK | 19 | 14 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE DN | 103 | 67 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 156 | 162 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 207 | 213 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA | ||||
| miR-124/506 | 207 | 213 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 70 | 76 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-133 | 109 | 115 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.