Gene Page: RGS14
Summary ?
| GeneID | 10636 |
| Symbol | RGS14 |
| Synonyms | - |
| Description | regulator of G-protein signaling 14 |
| Reference | MIM:602513|HGNC:HGNC:9996|Ensembl:ENSG00000169220|HPRD:03944|Vega:OTTHUMG00000163324 |
| Gene type | protein-coding |
| Map location | 5q35.3 |
| Pascal p-value | 0.12 |
| Sherlock p-value | 0.031 |
| Fetal beta | -0.9 |
| eGene | Cerebellum Frontal Cortex BA9 Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0036 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16865055 | chr2 | 224523383 | RGS14 | 10636 | 0.07 | trans | ||
| rs16922710 | chr10 | 23087297 | RGS14 | 10636 | 0.13 | trans | ||
| rs443222 | chr16 | 83889679 | RGS14 | 10636 | 0.08 | trans | ||
| rs824575 | chr16 | 83891459 | RGS14 | 10636 | 0.08 | trans | ||
| rs1883665 | chrX | 17650382 | RGS14 | 10636 | 0.12 | trans |
Section II. Transcriptome annotation
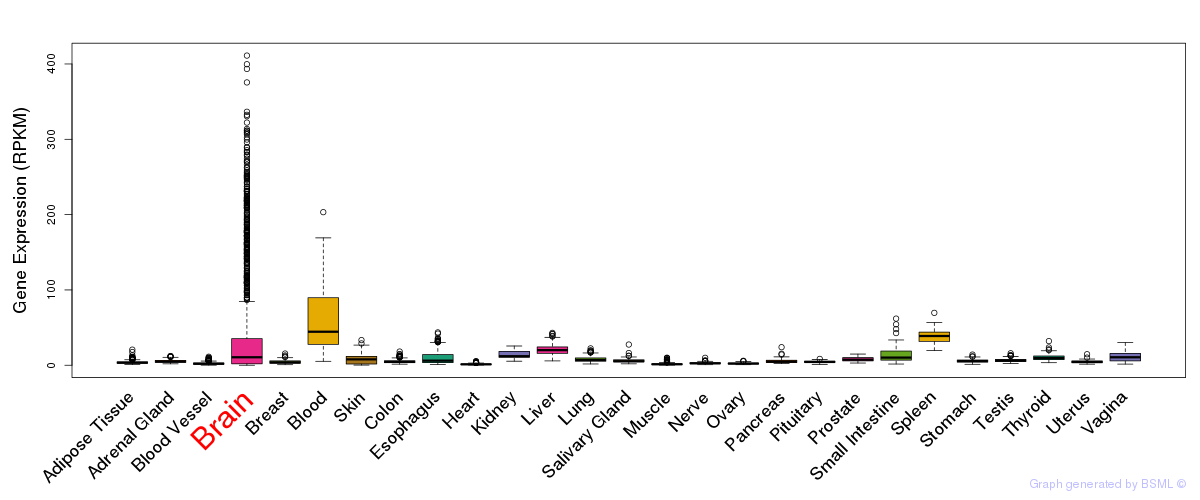
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC113191.1 | 0.93 | 0.90 |
| ZNF121 | 0.88 | 0.88 |
| PCDHB11 | 0.87 | 0.84 |
| ZNF852 | 0.85 | 0.82 |
| GATAD2B | 0.85 | 0.91 |
| IGF1R | 0.85 | 0.90 |
| RC3H1 | 0.84 | 0.85 |
| KIAA1853 | 0.84 | 0.83 |
| RAD54L2 | 0.84 | 0.92 |
| TMEM213 | 0.84 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.52 | -0.68 |
| C5orf53 | -0.50 | -0.69 |
| AF347015.31 | -0.49 | -0.79 |
| LHPP | -0.49 | -0.52 |
| HSD17B14 | -0.49 | -0.71 |
| AF347015.27 | -0.49 | -0.78 |
| ACOT13 | -0.48 | -0.61 |
| MT-CO2 | -0.48 | -0.78 |
| IFI27 | -0.48 | -0.76 |
| S100B | -0.48 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005057 | receptor signaling protein activity | IEA | - | |
| GO:0005096 | GTPase activator activity | TAS | 9168931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009968 | negative regulation of signal transduction | IEA | - | |
| GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway | TAS | 9168931 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP | 53 | 35 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K27ME3 | 70 | 35 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-324-3p | 294 | 300 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.