Gene Page: FTCD
Summary ?
| GeneID | 10841 |
| Symbol | FTCD |
| Synonyms | LCHC1 |
| Description | formimidoyltransferase cyclodeaminase |
| Reference | MIM:606806|HGNC:HGNC:3974|Ensembl:ENSG00000160282|HPRD:08430|Vega:OTTHUMG00000090488 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.198 |
| Fetal beta | -0.403 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7281816 | 21 | 47578569 | FTCD | ENSG00000160282.9 | 9.694E-7 | 0.01 | -3088 | gtex_brain_putamen_basal |
| rs73144711 | 21 | 47579051 | FTCD | ENSG00000160282.9 | 3.674E-7 | 0.01 | -3570 | gtex_brain_putamen_basal |
| rs56163044 | 21 | 47579752 | FTCD | ENSG00000160282.9 | 3.43E-7 | 0.01 | -4271 | gtex_brain_putamen_basal |
| rs55720515 | 21 | 47579901 | FTCD | ENSG00000160282.9 | 3.459E-7 | 0.01 | -4420 | gtex_brain_putamen_basal |
| rs7282697 | 21 | 47580415 | FTCD | ENSG00000160282.9 | 3.454E-7 | 0.01 | -4934 | gtex_brain_putamen_basal |
| rs55644041 | 21 | 47580532 | FTCD | ENSG00000160282.9 | 7.075E-7 | 0.01 | -5051 | gtex_brain_putamen_basal |
| rs56283108 | 21 | 47580575 | FTCD | ENSG00000160282.9 | 3.962E-7 | 0.01 | -5094 | gtex_brain_putamen_basal |
| rs56310297 | 21 | 47580600 | FTCD | ENSG00000160282.9 | 5.268E-7 | 0.01 | -5119 | gtex_brain_putamen_basal |
| rs75660802 | 21 | 47581074 | FTCD | ENSG00000160282.9 | 2.494E-7 | 0.01 | -5593 | gtex_brain_putamen_basal |
| rs113835421 | 21 | 47581693 | FTCD | ENSG00000160282.9 | 4.155E-7 | 0.01 | -6212 | gtex_brain_putamen_basal |
| rs58861127 | 21 | 47588735 | FTCD | ENSG00000160282.9 | 1.487E-7 | 0.01 | -13254 | gtex_brain_putamen_basal |
| rs8134429 | 21 | 47600157 | FTCD | ENSG00000160282.9 | 1.603E-7 | 0.01 | -24676 | gtex_brain_putamen_basal |
| rs79044044 | 21 | 47603849 | FTCD | ENSG00000160282.9 | 1.906E-6 | 0.01 | -28368 | gtex_brain_putamen_basal |
| rs73144744 | 21 | 47611285 | FTCD | ENSG00000160282.9 | 2.696E-7 | 0.01 | -35804 | gtex_brain_putamen_basal |
| rs9647243 | 21 | 47740415 | FTCD | ENSG00000160282.9 | 3.447E-7 | 0.01 | -164934 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
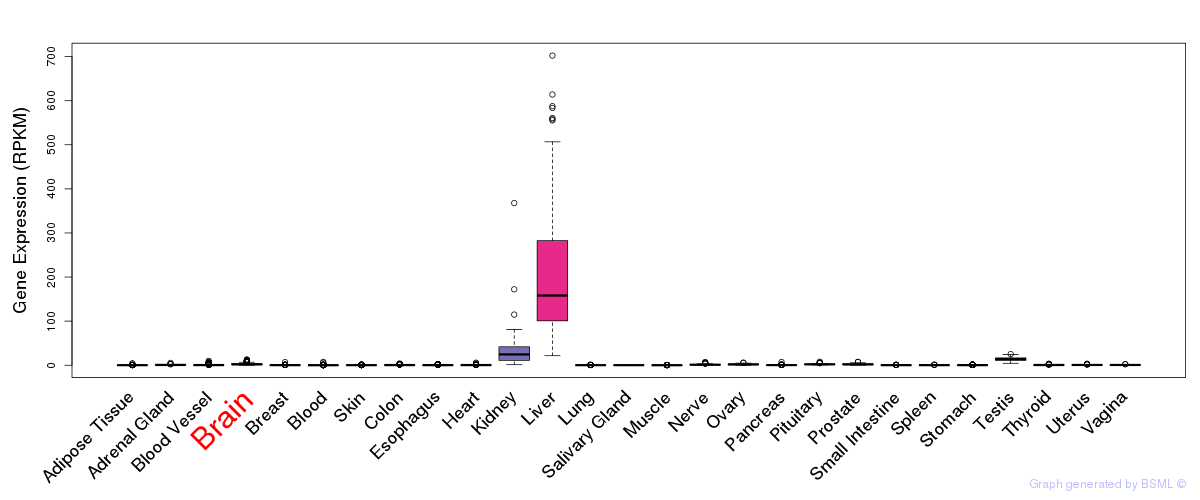
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRKACA | 0.91 | 0.90 |
| ENO2 | 0.91 | 0.92 |
| CLSTN3 | 0.91 | 0.92 |
| ABCG4 | 0.90 | 0.88 |
| ATP6AP1 | 0.90 | 0.93 |
| SV2A | 0.89 | 0.90 |
| AP2A1 | 0.89 | 0.90 |
| DCTN1 | 0.89 | 0.90 |
| PLBD2 | 0.88 | 0.87 |
| ATP1A3 | 0.88 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.54 | -0.48 |
| GNG11 | -0.50 | -0.51 |
| AP002478.3 | -0.49 | -0.50 |
| AF347015.31 | -0.48 | -0.43 |
| AF347015.2 | -0.48 | -0.37 |
| AF347015.8 | -0.47 | -0.42 |
| AF347015.18 | -0.47 | -0.42 |
| MT-CO2 | -0.47 | -0.43 |
| C1orf54 | -0.46 | -0.48 |
| NOSTRIN | -0.46 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030409 | glutamate formimidoyltransferase activity | IEA | glutamate (GO term level: 9) | - |
| GO:0005542 | folic acid binding | IEA | - | |
| GO:0016829 | lyase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0030412 | formimidoyltetrahydrofolate cyclodeaminase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006760 | folic acid and derivative metabolic process | TAS | 10029623 | |
| GO:0006547 | histidine metabolic process | IEA | - | |
| GO:0044237 | cellular metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 10029623 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HISTIDINE METABOLISM | 29 | 19 | All SZGR 2.0 genes in this pathway |
| KEGG ONE CARBON POOL BY FOLATE | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 21Q22 AMPLICON | 16 | 11 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 DN | 38 | 22 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |