Gene Page: TUBGCP2
Summary ?
| GeneID | 10844 |
| Symbol | TUBGCP2 |
| Synonyms | GCP-2|GCP2|Grip103|SPBC97|Spc97p|h103p|hGCP2|hSpc97 |
| Description | tubulin gamma complex associated protein 2 |
| Reference | HGNC:HGNC:18599|Ensembl:ENSG00000130640|HPRD:18245|Vega:OTTHUMG00000019319 |
| Gene type | protein-coding |
| Map location | 10q26.3 |
| Pascal p-value | 0.218 |
| Sherlock p-value | 0.998 |
| Fetal beta | 0.382 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03487 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10870786 | 10 | 135121752 | TUBGCP2 | 5.39E-10 | -0.025 | 8.9E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17795337 | chr6 | 136492658 | TUBGCP2 | 10844 | 0.05 | trans |
Section II. Transcriptome annotation
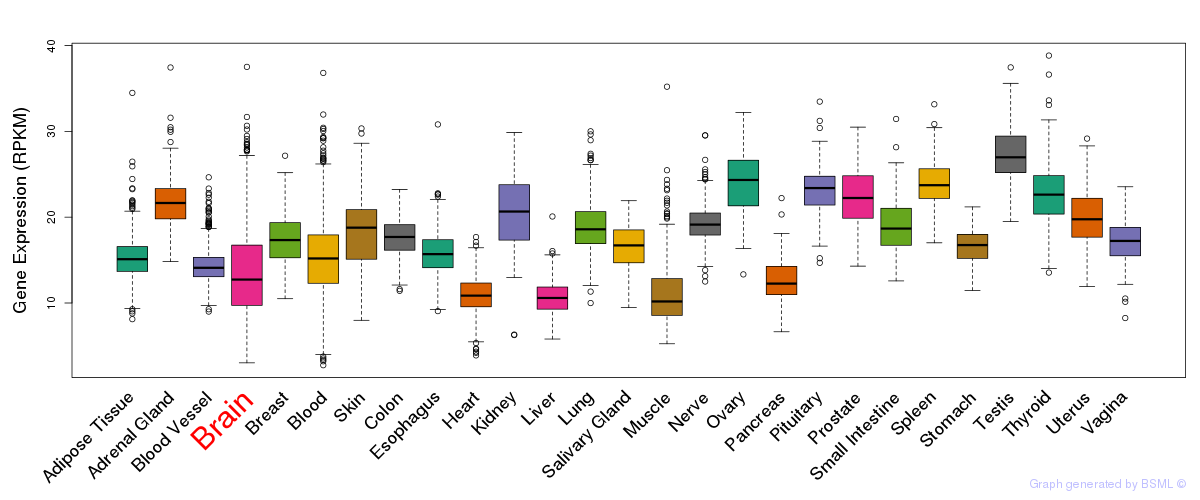
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000226 | microtubule cytoskeleton organization | IEA | - | |
| GO:0006461 | protein complex assembly | TAS | 9566967 | |
| GO:0007020 | microtubule nucleation | TAS | 9566967 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000922 | spindle pole | IEA | - | |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005815 | microtubule organizing center | IEA | - | |
| GO:0005881 | cytoplasmic microtubule | TAS | 9566967 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| WHITEHURST PACLITAXEL SENSITIVITY | 39 | 19 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |