Gene Page: COPS8
Summary ?
| GeneID | 10920 |
| Symbol | COPS8 |
| Synonyms | COP9|CSN8|SGN8 |
| Description | COP9 signalosome subunit 8 |
| Reference | MIM:616011|HGNC:HGNC:24335|HPRD:16737| |
| Gene type | protein-coding |
| Map location | 2q37.3 |
| Pascal p-value | 0.825 |
| Sherlock p-value | 0.632 |
| DMG | 1 (# studies) |
| eGene | Hippocampus |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19102858 | 2 | 237993901 | COPS8 | -0.02 | 0.62 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
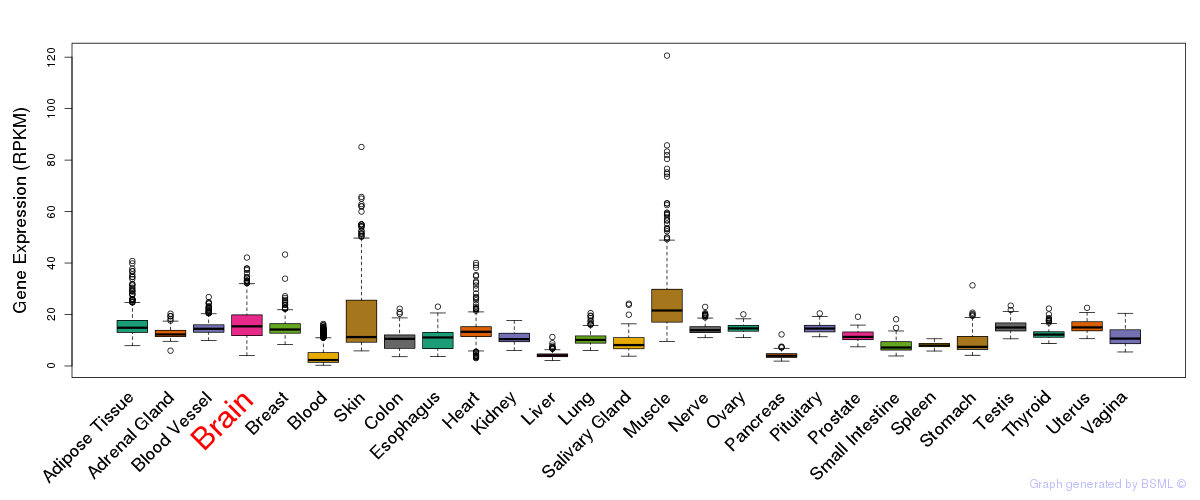
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLXNA3 | 0.94 | 0.88 |
| DCHS1 | 0.93 | 0.94 |
| KDM6B | 0.92 | 0.89 |
| PLEKHG2 | 0.92 | 0.93 |
| AD000671.1 | 0.92 | 0.84 |
| NINL | 0.92 | 0.91 |
| KIAA1305 | 0.91 | 0.91 |
| C20orf117 | 0.91 | 0.83 |
| BCL9L | 0.91 | 0.86 |
| MLXIP | 0.91 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.66 | -0.77 |
| HLA-F | -0.66 | -0.66 |
| ACOT13 | -0.66 | -0.78 |
| FBXO2 | -0.65 | -0.66 |
| ALDOC | -0.65 | -0.63 |
| FAM162A | -0.63 | -0.75 |
| ACYP2 | -0.63 | -0.72 |
| CA4 | -0.63 | -0.73 |
| AIFM3 | -0.63 | -0.59 |
| LDHD | -0.62 | -0.59 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | CSN8 interacts with CSN3. | BIND | 12615944 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | CSN4 interacts with CSN8. | BIND | 12615944 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | - | HPRD,BioGRID | 9707402 |14647295 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS7A | MGC110877 | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS7B | FLJ12612 | MGC111077 | COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | CSN8 interacts with itself. | BIND | 12615944 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | - | HPRD | 12220626 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | Co-purification | BioGRID | 9707402 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | - | HPRD | 11337588 |
| ITPK1 | ITRPK1 | inositol 1,3,4-triphosphate 5/6 kinase | - | HPRD | 12324474 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| BARRIER COLON CANCER RECURRENCE UP | 42 | 28 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA UP | 16 | 7 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |