Gene Page: SLC27A5
Summary ?
| GeneID | 10998 |
| Symbol | SLC27A5 |
| Synonyms | ACSB|ACSVL6|BACS|BAL|FACVL3|FATP-5|FATP5|VLACSR|VLCS-H2|VLCSH2 |
| Description | solute carrier family 27 member 5 |
| Reference | MIM:603314|HGNC:HGNC:10999|Ensembl:ENSG00000083807|HPRD:04499|Vega:OTTHUMG00000183543 |
| Gene type | protein-coding |
| Map location | 19q13.43 |
| Pascal p-value | 0.03 |
| Sherlock p-value | 0.038 |
| Fetal beta | -0.307 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
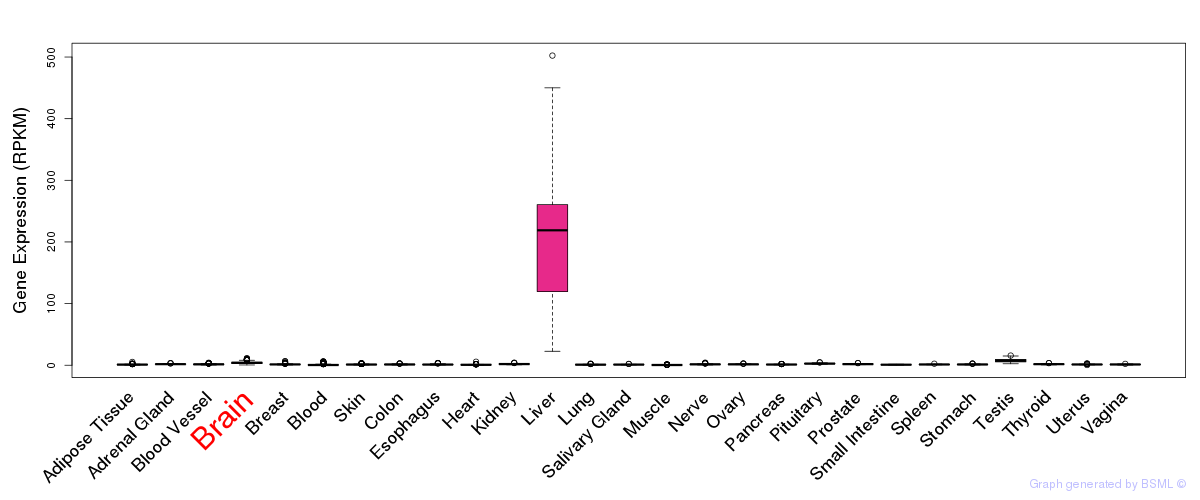
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBCA | 0.85 | 0.87 |
| PPIH | 0.84 | 0.83 |
| SARNP | 0.83 | 0.83 |
| CFDP1 | 0.83 | 0.80 |
| HYPK | 0.83 | 0.78 |
| DNAJC8 | 0.82 | 0.81 |
| SUMO2 | 0.81 | 0.83 |
| PFDN2 | 0.81 | 0.81 |
| CXorf26 | 0.80 | 0.79 |
| LSM8 | 0.80 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TINAGL1 | -0.60 | -0.71 |
| HLA-F | -0.60 | -0.67 |
| HLA-E | -0.59 | -0.69 |
| GPER | -0.58 | -0.68 |
| SLC6A12 | -0.58 | -0.68 |
| FGR | -0.57 | -0.67 |
| ATP10A | -0.57 | -0.67 |
| AF347015.9 | -0.56 | -0.67 |
| AF347015.2 | -0.55 | -0.66 |
| PRELP | -0.55 | -0.64 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PRIMARY BILE ACID BIOSYNTHESIS | 16 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME BILE ACID AND BILE SALT METABOLISM | 27 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | 15 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | 10 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | 19 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS DN | 47 | 33 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 DN | 65 | 39 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |