Gene Page: CHD4
Summary ?
| GeneID | 1108 |
| Symbol | CHD4 |
| Synonyms | CHD-4|Mi-2b|Mi2-BETA |
| Description | chromodomain helicase DNA binding protein 4 |
| Reference | MIM:603277|HGNC:HGNC:1919|Ensembl:ENSG00000111642|HPRD:04472|Vega:OTTHUMG00000169164 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.803 |
| Sherlock p-value | 0.478 |
| TADA p-value | 0.026 |
| Fetal beta | 0.178 |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Girard_2011 | Whole Exome Sequencing analysis | The data set included 15 DNMs found from the exomes of 14 schizophrenia probands and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHD4 | chr12 | 6707226 | G | A | NM_001273 | p.576R>W | missense | Schizophrenia | DNM:Girard_2011 |
Section II. Transcriptome annotation
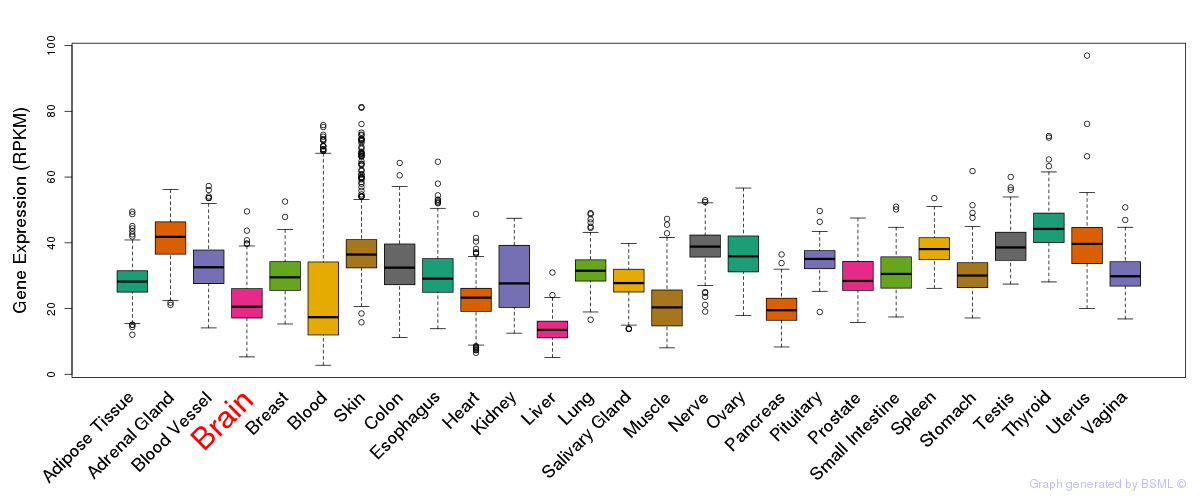
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MX1 | 0.82 | 0.82 |
| TAPBPL | 0.80 | 0.85 |
| EPHX1 | 0.80 | 0.80 |
| TMBIM1 | 0.79 | 0.87 |
| TNFSF13 | 0.79 | 0.86 |
| CYP11A1 | 0.79 | 0.81 |
| TMPRSS3 | 0.78 | 0.73 |
| SLC14A1 | 0.78 | 0.77 |
| GJB6 | 0.77 | 0.85 |
| AGXT2L1 | 0.77 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF14 | -0.49 | -0.76 |
| NKIRAS2 | -0.49 | -0.71 |
| RTF1 | -0.49 | -0.71 |
| CRMP1 | -0.48 | -0.72 |
| PCDHB18 | -0.48 | -0.73 |
| ZWILCH | -0.48 | -0.69 |
| PJA1 | -0.48 | -0.69 |
| DPF1 | -0.48 | -0.71 |
| ZNF821 | -0.48 | -0.71 |
| NOL4 | -0.48 | -0.70 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | - | HPRD,BioGRID | 10545197 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 9804427 |10220385 |12920132 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 9804427 |10545197 |12493763 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF2 | HELIOS | MGC34330 | ZNF1A2 | ZNFN1A2 | IKAROS family zinc finger 2 (Helios) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF3 | AIO | AIOLOS | ZNFN1A3 | IKAROS family zinc finger 3 (Aiolos) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF4 | EOS | KIAA1782 | ZNFN1A4 | IKAROS family zinc finger 4 (Eos) | - | HPRD,BioGRID | 12015313 |
| MBD3 | - | methyl-CpG binding domain protein 3 | Affinity Capture-Western | BioGRID | 12354758 |
| MTA1 | - | metastasis associated 1 | Affinity Capture-MS | BioGRID | 12920132 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12920132 |
| SATB1 | - | SATB homeobox 1 | Affinity Capture-Western Co-fractionation | BioGRID | 12374985 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | - | HPRD | 14530259 |
| XRN1 | DKFZp434P0721 | DKFZp686B22225 | DKFZp686F19113 | FLJ41903 | SEP1 | 5'-3' exoribonuclease 1 | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG UP | 44 | 29 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 3 DN | 10 | 7 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX1 UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN DAUNORUBICIN B ALL UP | 10 | 9 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN DAUNORUBICIN ALL UP | 7 | 5 | All SZGR 2.0 genes in this pathway |