Gene Page: POLI
Summary ?
| GeneID | 11201 |
| Symbol | POLI |
| Synonyms | RAD30B|RAD3OB |
| Description | polymerase (DNA) iota |
| Reference | MIM:605252|HGNC:HGNC:9182|Ensembl:ENSG00000101751|HPRD:16094|Vega:OTTHUMG00000132704 |
| Gene type | protein-coding |
| Map location | 18q21.1 |
| Pascal p-value | 0.902 |
| Fetal beta | 1.03 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | POLI | 11201 | 0.16 | trans | ||
| rs2085670 | 18 | 51767894 | POLI | ENSG00000101751.6 | 3.153E-6 | 0.01 | -27880 | gtex_brain_putamen_basal |
| rs6508285 | 18 | 51769949 | POLI | ENSG00000101751.6 | 2.834E-6 | 0.01 | -25825 | gtex_brain_putamen_basal |
| rs8089411 | 18 | 51771322 | POLI | ENSG00000101751.6 | 3.826E-6 | 0.01 | -24452 | gtex_brain_putamen_basal |
| rs8093601 | 18 | 51772473 | POLI | ENSG00000101751.6 | 1.688E-6 | 0.01 | -23301 | gtex_brain_putamen_basal |
| rs11083046 | 18 | 51781019 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | -14755 | gtex_brain_putamen_basal |
| rs11083047 | 18 | 51781894 | POLI | ENSG00000101751.6 | 3.725E-6 | 0.01 | -13880 | gtex_brain_putamen_basal |
| rs10048321 | 18 | 51782775 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | -12999 | gtex_brain_putamen_basal |
| rs55671046 | 18 | 51784064 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | -11710 | gtex_brain_putamen_basal |
| rs28393808 | 18 | 51787992 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | -7782 | gtex_brain_putamen_basal |
| rs649420 | 18 | 51793690 | POLI | ENSG00000101751.6 | 4.374E-6 | 0.01 | -2084 | gtex_brain_putamen_basal |
| rs36047427 | 18 | 51793968 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | -1806 | gtex_brain_putamen_basal |
| rs3730668 | 18 | 51795847 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | 73 | gtex_brain_putamen_basal |
| rs2276182 | 18 | 51798047 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | 2273 | gtex_brain_putamen_basal |
| rs3730714 | 18 | 51802708 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | 6934 | gtex_brain_putamen_basal |
| rs3730716 | 18 | 51802787 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | 7013 | gtex_brain_putamen_basal |
| rs3730775 | 18 | 51813966 | POLI | ENSG00000101751.6 | 5.261E-7 | 0.01 | 18192 | gtex_brain_putamen_basal |
| rs3730783 | 18 | 51815283 | POLI | ENSG00000101751.6 | 5.182E-7 | 0.01 | 19509 | gtex_brain_putamen_basal |
| rs1903141 | 18 | 51817587 | POLI | ENSG00000101751.6 | 1.099E-6 | 0.01 | 21813 | gtex_brain_putamen_basal |
| rs34162186 | 18 | 51825773 | POLI | ENSG00000101751.6 | 5.305E-7 | 0.01 | 29999 | gtex_brain_putamen_basal |
| rs1893958 | 18 | 51829629 | POLI | ENSG00000101751.6 | 1.117E-6 | 0.01 | 33855 | gtex_brain_putamen_basal |
| rs138245320 | 18 | 51831039 | POLI | ENSG00000101751.6 | 3.969E-6 | 0.01 | 35265 | gtex_brain_putamen_basal |
| rs34163044 | 18 | 51851616 | POLI | ENSG00000101751.6 | 1.692E-6 | 0.01 | 55842 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
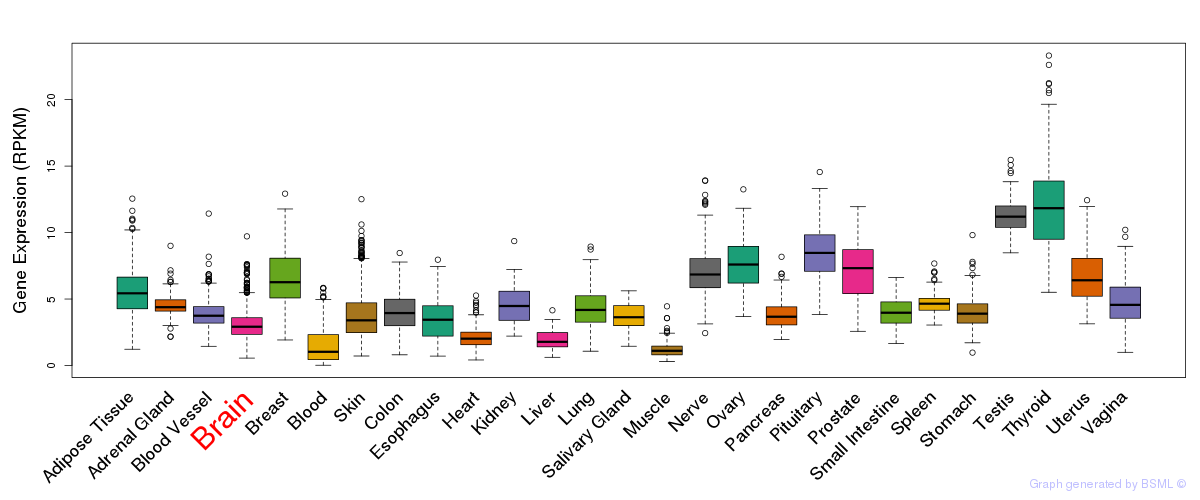
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |