Gene Page: EGLN2
Summary ?
| GeneID | 112398 |
| Symbol | EGLN2 |
| Synonyms | EIT6|HIF-PH1|HIFPH1|HPH-1|HPH-3|PHD1 |
| Description | egl-9 family hypoxia-inducible factor 2 |
| Reference | MIM:606424|HGNC:HGNC:14660|Ensembl:ENSG00000269858|HPRD:06970|Vega:OTTHUMG00000182703 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.279 |
| Sherlock p-value | 0.219 |
| Fetal beta | 0.489 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08080060 | 19 | 41307090 | EGLN2 | 1.639E-4 | 0.349 | 0.033 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16894557 | chr6 | 28999825 | EGLN2 | 112398 | 0.06 | trans | ||
| rs17795337 | chr6 | 136492658 | EGLN2 | 112398 | 0.14 | trans |
Section II. Transcriptome annotation
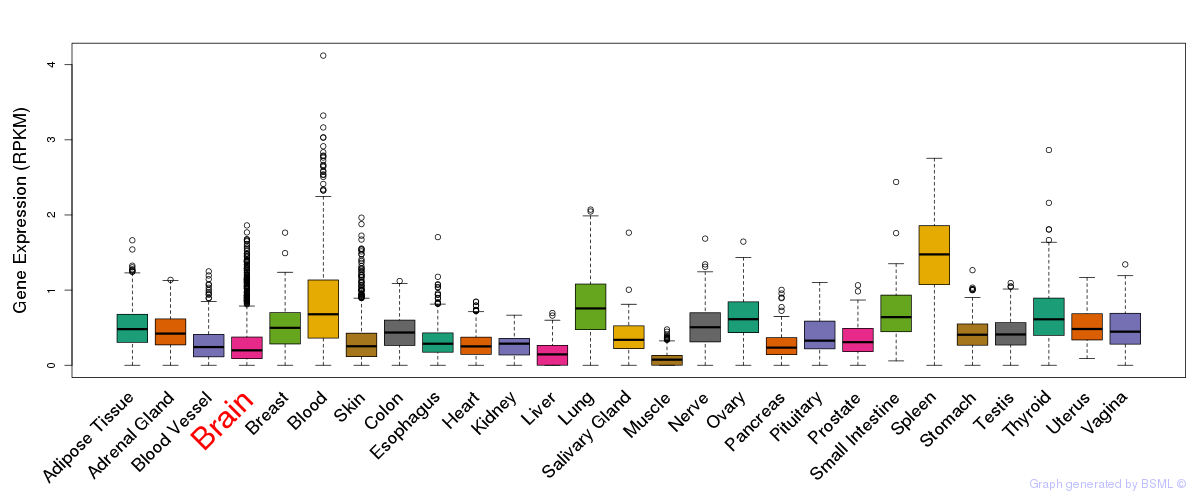
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | 18 | 12 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |