Gene Page: ECD
Summary ?
| GeneID | 11319 |
| Symbol | ECD |
| Synonyms | GCR2|HSGT1|SGT1 |
| Description | ecdysoneless cell cycle regulator |
| Reference | MIM:616464|HGNC:HGNC:17029|Ensembl:ENSG00000122882|HPRD:11032|Vega:OTTHUMG00000018451 |
| Gene type | protein-coding |
| Map location | 10q22.2 |
| Pascal p-value | 0.604 |
| eGene | Cerebellar Hemisphere Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08879241 | 10 | 74927972 | FAM149B1;ECD | 4.38E-5 | -0.433 | 0.021 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6749607 | chr2 | 143944077 | ECD | 11319 | 0 | trans | ||
| rs6743877 | chr2 | 143954689 | ECD | 11319 | 0.01 | trans | ||
| rs6728893 | chr2 | 143966610 | ECD | 11319 | 0.01 | trans | ||
| rs6430007 | chr2 | 143971325 | ECD | 11319 | 0.01 | trans | ||
| rs13032052 | chr2 | 143976479 | ECD | 11319 | 0.04 | trans | ||
| rs17812607 | chr2 | 143980968 | ECD | 11319 | 0 | trans | ||
| rs7597991 | chr2 | 143981009 | ECD | 11319 | 0 | trans | ||
| rs13004383 | chr2 | 143981602 | ECD | 11319 | 0.01 | trans | ||
| rs10496945 | chr2 | 143998943 | ECD | 11319 | 0 | trans | ||
| rs13035017 | chr2 | 143999478 | ECD | 11319 | 0 | trans | ||
| rs9677486 | chr2 | 144021777 | ECD | 11319 | 0.11 | trans | ||
| rs16858944 | chr2 | 144022953 | ECD | 11319 | 0.14 | trans |
Section II. Transcriptome annotation
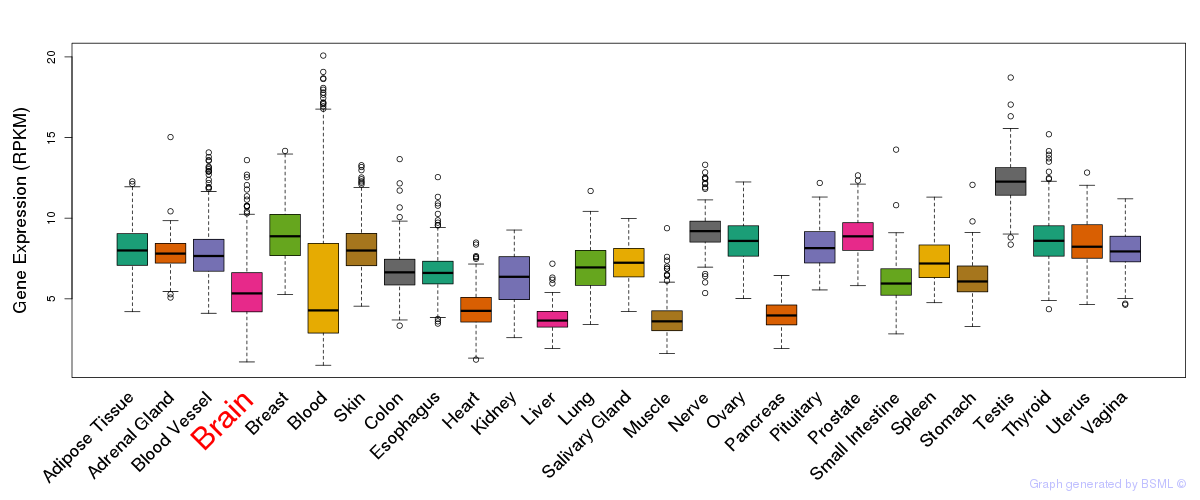
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |