Gene Page: TPH2
Summary ?
| GeneID | 121278 |
| Symbol | TPH2 |
| Synonyms | ADHD7|NTPH |
| Description | tryptophan hydroxylase 2 |
| Reference | MIM:607478|HGNC:HGNC:20692|Ensembl:ENSG00000139287|HPRD:09594|Vega:OTTHUMG00000169677 |
| Gene type | protein-coding |
| Map location | 12q21.1 |
| Pascal p-value | 0.016 |
| Fetal beta | -0.181 |
| Support | NEUROTRANSMITTER METABOLISM SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
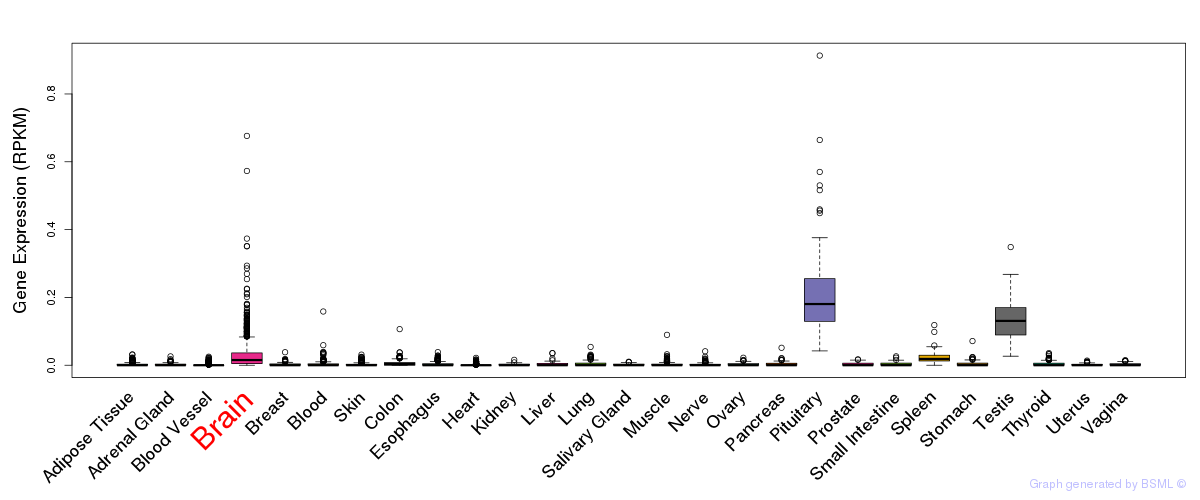
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0004497 | monooxygenase activity | IEA | - | |
| GO:0004510 | tryptophan 5-monooxygenase activity | IEA | - | |
| GO:0016597 | amino acid binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006587 | serotonin biosynthetic process from tryptophan | IEA | serotonin, Neurotransmitter (GO term level: 10) | - |
| GO:0042427 | serotonin biosynthetic process | IEA | serotonin, Neurotransmitter (GO term level: 9) | - |
| GO:0009072 | aromatic amino acid family metabolic process | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE DERIVED HORMONES | 15 | 12 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |