Gene Page: DEGS2
Summary ?
| GeneID | 123099 |
| Symbol | DEGS2 |
| Synonyms | C14orf66|DES2|FADS8 |
| Description | delta(4)-desaturase, sphingolipid 2 |
| Reference | MIM:610862|HGNC:HGNC:20113|HPRD:13134| |
| Gene type | protein-coding |
| Map location | 14q32.2 |
| Pascal p-value | 0.275 |
| Fetal beta | -1.063 |
| eGene | Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | DEGS2 | 123099 | 0.06 | trans | ||
| rs2481653 | chr1 | 176044120 | DEGS2 | 123099 | 0.05 | trans | ||
| rs2502827 | chr1 | 176044216 | DEGS2 | 123099 | 0.02 | trans | ||
| rs12405921 | chr1 | 206695730 | DEGS2 | 123099 | 0.12 | trans | ||
| rs16829545 | chr2 | 151977407 | DEGS2 | 123099 | 2.79E-12 | trans | ||
| rs3845734 | chr2 | 171125572 | DEGS2 | 123099 | 3.517E-5 | trans | ||
| rs7584986 | chr2 | 184111432 | DEGS2 | 123099 | 2.362E-6 | trans | ||
| rs6821203 | chr4 | 189475342 | DEGS2 | 123099 | 0.05 | trans | ||
| rs3111196 | chr5 | 54402889 | DEGS2 | 123099 | 0.04 | trans | ||
| rs17762315 | chr5 | 76807576 | DEGS2 | 123099 | 3.76E-4 | trans | ||
| rs6887062 | chr5 | 123850762 | DEGS2 | 123099 | 0.16 | trans | ||
| rs1368303 | chr5 | 147672388 | DEGS2 | 123099 | 0.08 | trans | ||
| rs2725663 | chr8 | 5518325 | DEGS2 | 123099 | 0.15 | trans | ||
| rs12281901 | chr11 | 83839991 | DEGS2 | 123099 | 0.05 | trans | ||
| rs647575 | chr13 | 52473715 | DEGS2 | 123099 | 0.1 | trans | ||
| rs16955618 | chr15 | 29937543 | DEGS2 | 123099 | 8.647E-17 | trans | ||
| rs1041786 | chr21 | 22617710 | DEGS2 | 123099 | 0.02 | trans |
Section II. Transcriptome annotation
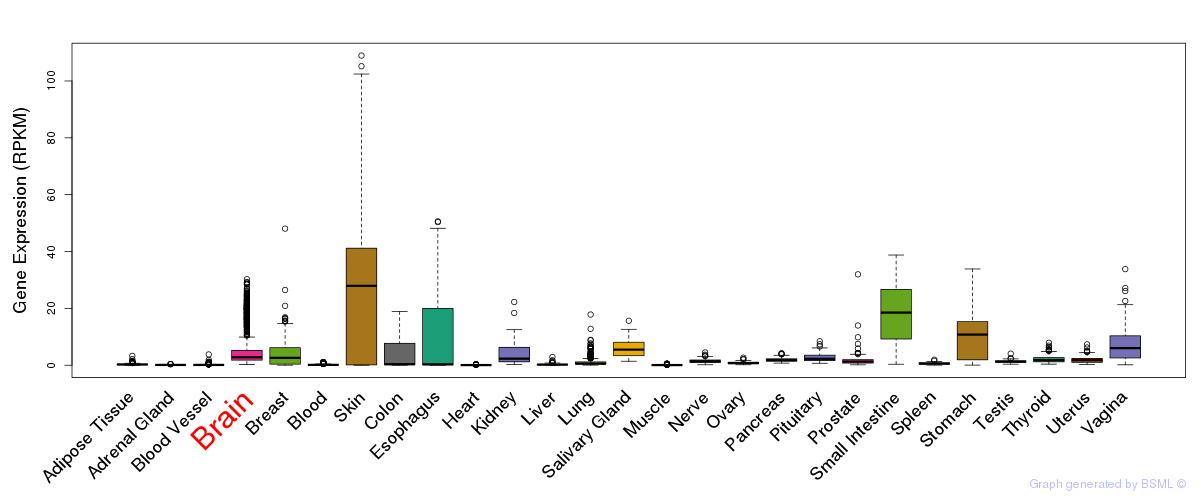
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPHINGOLIPID METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | 31 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID METABOLISM | 69 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |