Gene Page: CREB1
Summary ?
| GeneID | 1385 |
| Symbol | CREB1 |
| Synonyms | CREB|CREB-1 |
| Description | cAMP responsive element binding protein 1 |
| Reference | MIM:123810|HGNC:HGNC:2345|Ensembl:ENSG00000118260|HPRD:00442|Vega:OTTHUMG00000132936 |
| Gene type | protein-coding |
| Map location | 2q34 |
| Pascal p-value | 3.592E-6 |
| Sherlock p-value | 0.153 |
| Fetal beta | 1.69 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0239 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
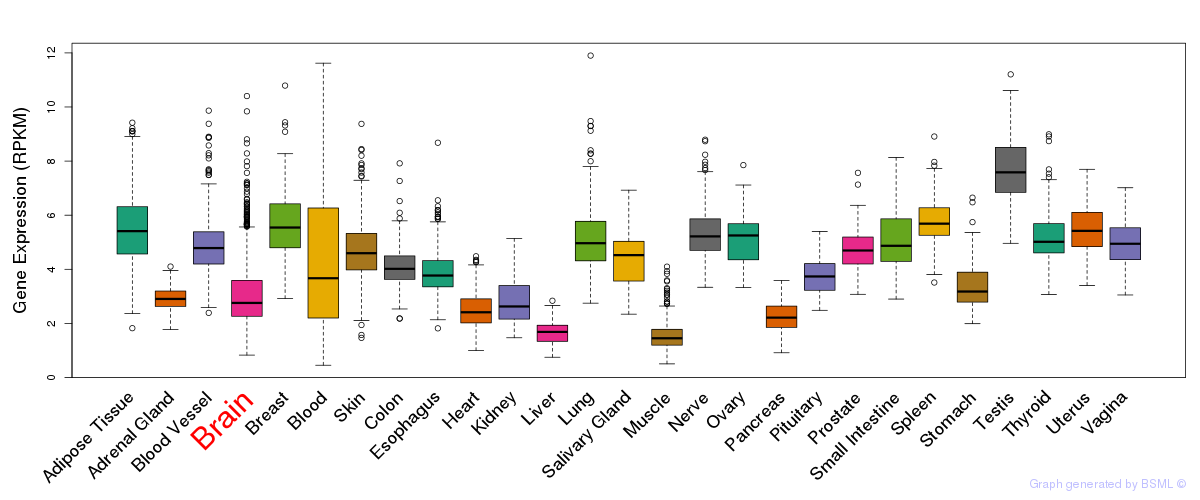
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDRG4 | 0.81 | 0.78 |
| GPR123 | 0.81 | 0.82 |
| SYNGR3 | 0.80 | 0.83 |
| GPR61 | 0.79 | 0.77 |
| C19orf28 | 0.78 | 0.79 |
| ZDHHC14 | 0.78 | 0.77 |
| SLC25A22 | 0.78 | 0.82 |
| PIGZ | 0.77 | 0.78 |
| PPAPDC3 | 0.77 | 0.80 |
| CLSTN3 | 0.77 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB13 | -0.41 | -0.49 |
| GTF3C6 | -0.40 | -0.39 |
| NSBP1 | -0.39 | -0.44 |
| ANP32B | -0.39 | -0.48 |
| EIF5B | -0.39 | -0.45 |
| RBMX2 | -0.38 | -0.39 |
| C21orf57 | -0.38 | -0.37 |
| GSTM2 | -0.37 | -0.38 |
| CWF19L2 | -0.36 | -0.41 |
| C9orf46 | -0.36 | -0.36 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 10909971 | |
| GO:0003712 | transcription cofactor activity | TAS | 8552098 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| GO:0046983 | protein dimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IDA | 8798441 | |
| GO:0007165 | signal transduction | TAS | 2974179 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 10909971 | |
| GO:0005654 | nucleoplasm | EXP | 9687510 |9770464 |9829964 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Reconstituted Complex | BioGRID | 7565761 |
| ATF1 | EWS-ATF1 | FUS/ATF-1 | TREB36 | activating transcription factor 1 | - | HPRD,BioGRID | 1655749 |8384217 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Affinity Capture-Western | BioGRID | 15073328 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Phenotypic Enhancement | BioGRID | 10945975 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | Biochemical Activity | BioGRID | 8663317 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | CREB interacts with CRE. | BIND | 15897899 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 12773552 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CREB1 (CREB) interacts with CREBBP (CBP). | BIND | 16007092 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with CREB. | BIND | 15454081 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 9413984 |
| CREM | ICER | MGC111110 | MGC17881 | MGC41893 | hCREM-2 | cAMP responsive element modulator | - | HPRD | 7809053 |
| CRTC1 | FLJ14027 | KIAA0616 | MECT1 | TORC1 | WAMTP1 | CREB regulated transcription coactivator 1 | TORC1 interacts with CREB. | BIND | 15454081 |
| CYP19A1 | ARO | ARO1 | CPV1 | CYAR | CYP19 | MGC104309 | P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 | CREB1 (CREB) interacts with the CYP19A1 (aromatase PII) promoter. | BIND | 15688015 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | DYRK1A interacts with and phosphorylates CREB. This interaction was modeled on a demonstrated interaction between human DYRK1A and CREB from rat. | BIND | 15694837 |
| EDF1 | EDF-1 | MBF1 | MGC9058 | endothelial differentiation-related factor 1 | - | HPRD,BioGRID | 10567391 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 10506141 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | Affinity Capture-Western Two-hybrid | BioGRID | 11046156 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | Affinity Capture-Western Two-hybrid | BioGRID | 11046156 |
| FHL5 | ACT | FLJ33049 | KIAA0776 | RP3-393D12.2 | dJ393D12.2 | four and a half LIM domains 5 | - | HPRD,BioGRID | 11046156 |
| FOSL1 | FRA | FRA1 | fra-1 | FOS-like antigen 1 | CREB interacts with the region of the FRA-1 promoter containing the SRE and ATF sites. | BIND | 15806162 |
| GLI2 | HPE9 | THP1 | THP2 | GLI-Kruppel family member GLI2 | - | HPRD,BioGRID | 10074179 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD | 8628277 |
| HNF1B | FJHN | HNF1beta | HNF2 | HPC11 | LF-B3 | LFB3 | MODY5 | TCF2 | VHNF1 | HNF1 homeobox B | Two-hybrid | BioGRID | 9671480 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD,BioGRID | 10720489 |
| KCNIP3 | CSEN | DREAM | KCHIP3 | MGC18289 | Kv channel interacting protein 3, calsenilin | - | HPRD,BioGRID | 12198160 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD,BioGRID | 11377386 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 7621901 |8449898 |
| NR4A2 | HZF-3 | NOT | NURR1 | RNR1 | TINUR | nuclear receptor subfamily 4, group A, member 2 | An unspecified isoform of CREB binds to Nurr1 promoter. | BIND | 15961999 |
| PDX1 | GSF | IDX-1 | IPF1 | IUF1 | MODY4 | PDX-1 | STF-1 | pancreatic and duodenal homeobox 1 | - | HPRD | 7505393 |
| PRKACA | MGC102831 | MGC48865 | PKACA | protein kinase, cAMP-dependent, catalytic, alpha | Reconstituted Complex | BioGRID | 11500364 |
| RAB1A | DKFZp564B163 | RAB1 | YPT1 | RAB1A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Two-hybrid | BioGRID | 10866654 |
| RECQL5 | FLJ90603 | RECQ5 | RecQ protein-like 5 | Two-hybrid | BioGRID | 16169070 |
| RPS6KA4 | MSK2 | RSK-B | ribosomal protein S6 kinase, 90kDa, polypeptide 4 | - | HPRD,BioGRID | 9792677 |
| RPS6KA5 | MGC1911 | MSK1 | MSPK1 | RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | Biochemical Activity Reconstituted Complex | BioGRID | 9687510 |11500364 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | - | HPRD | 12805224 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10848610 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | YY1 interacts with CREB. | BIND | 7769693 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CACAM PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HCMV PATHWAY | 17 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARENRF2 PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK5 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM1 PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA S PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID LPA4 PATHWAY | 15 | 10 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID AR NONGENOMIC PATHWAY | 31 | 27 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID LYMPH ANGIOGENESIS PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK7 TARGETS | 8 | 8 | All SZGR 2.0 genes in this pathway |
| HAEGERSTRAND RESPONSE TO IMATINIB | 9 | 5 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM6 | 46 | 32 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 362 | 368 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 362 | 368 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 164 | 170 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-181 | 168 | 174 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 455 | 462 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-203.1 | 89 | 95 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-25/32/92/363/367 | 435 | 441 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 164 | 171 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-323 | 402 | 408 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-34b | 114 | 120 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-450 | 108 | 114 | m8 | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-500 | 433 | 440 | 1A,m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-505 | 448 | 454 | m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-96 | 456 | 462 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.