Gene Page: AIFM3
Summary ?
| GeneID | 150209 |
| Symbol | AIFM3 |
| Synonyms | AIFL |
| Description | apoptosis inducing factor, mitochondria associated 3 |
| Reference | HGNC:HGNC:26398|Ensembl:ENSG00000183773|HPRD:08085|Vega:OTTHUMG00000150804 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.312 |
| Sherlock p-value | 0.208 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11582961 | chr1 | 184023058 | AIFM3 | 150209 | 0.06 | trans |
Section II. Transcriptome annotation
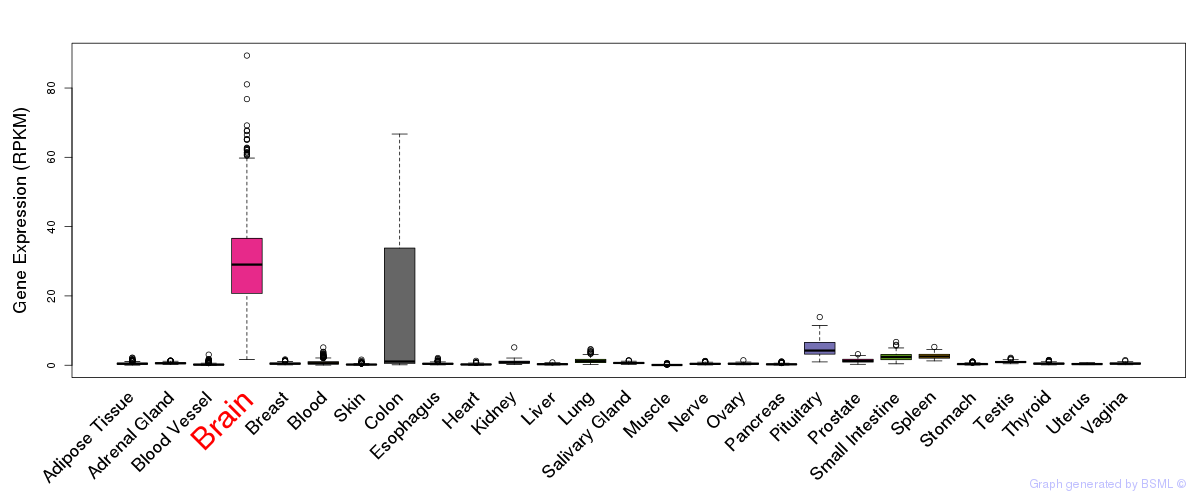
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0008656 | caspase activator activity | IDA | 15764604 | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050660 | FAD binding | IEA | - | |
| GO:0051537 | 2 iron, 2 sulfur cluster binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006917 | induction of apoptosis | IDA | 15764604 | |
| GO:0008635 | caspase activation via cytochrome c | IDA | 15764604 | |
| GO:0006810 | transport | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0022900 | electron transport chain | IEA | - | |
| GO:0045454 | cell redox homeostasis | IEA | - | |
| GO:0051882 | mitochondrial depolarization | IDA | 15764604 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IDA | 15764604 | |
| GO:0005634 | nucleus | IDA | 15764604 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IDA | 15764604 | |
| GO:0005743 | mitochondrial inner membrane | IDA | 15764604 | |
| GO:0005783 | endoplasmic reticulum | IDA | 15764604 | |
| GO:0005886 | plasma membrane | IDA | 15764604 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONG INTERACT WITH PTTG1 | 57 | 32 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-96 | 68 | 74 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.