Gene Page: CYP27A1
Summary ?
| GeneID | 1593 |
| Symbol | CYP27A1 |
| Synonyms | CP27|CTX|CYP27 |
| Description | cytochrome P450 family 27 subfamily A member 1 |
| Reference | MIM:606530|HGNC:HGNC:2605|Ensembl:ENSG00000135929|HPRD:05939|Vega:OTTHUMG00000048238 |
| Gene type | protein-coding |
| Map location | 2q35 |
| Pascal p-value | 0.991 |
| Sherlock p-value | 0.183 |
| Fetal beta | -1.264 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11656351 | chr17 | 9489574 | CYP27A1 | 1593 | 0.03 | trans | ||
| rs4501739 | chrX | 35960848 | CYP27A1 | 1593 | 0.15 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
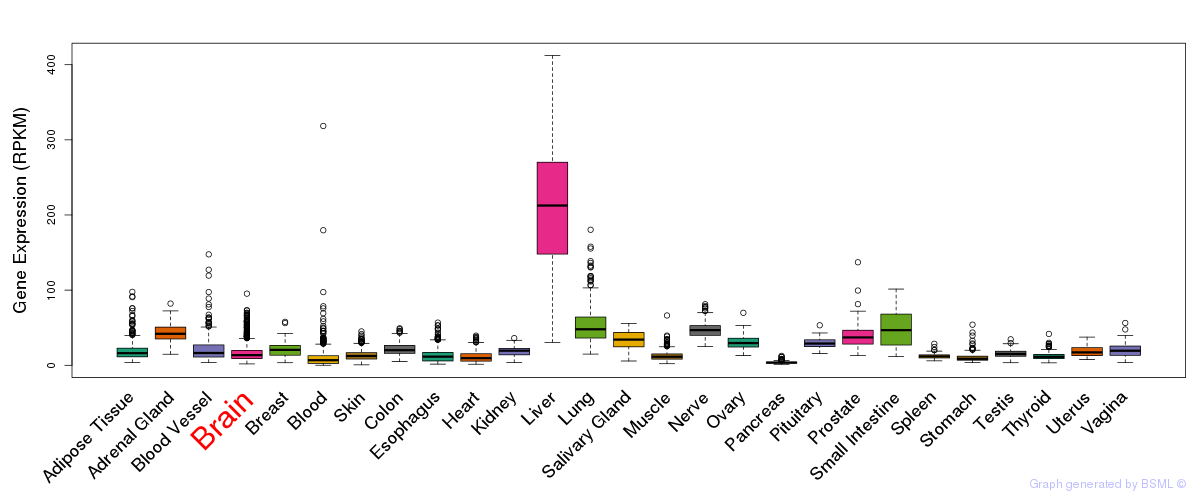
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| POFUT1 | 0.90 | 0.90 |
| NCAN | 0.89 | 0.90 |
| ANKFY1 | 0.88 | 0.89 |
| SEPN1 | 0.87 | 0.89 |
| SEC61A1 | 0.86 | 0.86 |
| MAPK1IP1L | 0.86 | 0.85 |
| SALL2 | 0.86 | 0.86 |
| NLGN3 | 0.85 | 0.89 |
| PCDHGB8P | 0.85 | 0.89 |
| CYFIP1 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.62 | -0.75 |
| MT-CO2 | -0.60 | -0.68 |
| C5orf53 | -0.60 | -0.63 |
| AF347015.31 | -0.60 | -0.67 |
| AF347015.27 | -0.58 | -0.65 |
| MYL3 | -0.57 | -0.60 |
| C1orf54 | -0.56 | -0.66 |
| AF347015.33 | -0.55 | -0.61 |
| AF347015.8 | -0.55 | -0.64 |
| FXYD1 | -0.55 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0020037 | heme binding | IEA | - | |
| GO:0008395 | steroid hydroxylase activity | EXP | 1708392 | |
| GO:0008395 | steroid hydroxylase activity | TAS | 9790667 | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0047749 | cholestanetriol 26-monooxygenase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005743 | mitochondrial inner membrane | IEA | - | |
| GO:0005759 | mitochondrial matrix | EXP | 1708392 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PRIMARY BILE ACID BIOSYNTHESIS | 16 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME BILE ACID AND BILE SALT METABOLISM | 27 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | 15 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | 10 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | 19 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOGENOUS STEROLS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS DN | 32 | 27 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |