Gene Page: DAXX
Summary ?
| GeneID | 1616 |
| Symbol | DAXX |
| Synonyms | BING2|DAP6|EAP1 |
| Description | death-domain associated protein |
| Reference | MIM:603186|HGNC:HGNC:2681|Ensembl:ENSG00000204209|HPRD:04424|Vega:OTTHUMG00000031203 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 2.567E-8 |
| Sherlock p-value | 0.215 |
| Fetal beta | -0.138 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18187593 | 6 | 33290972 | DAXX | 4.586E-4 | -0.375 | 0.046 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
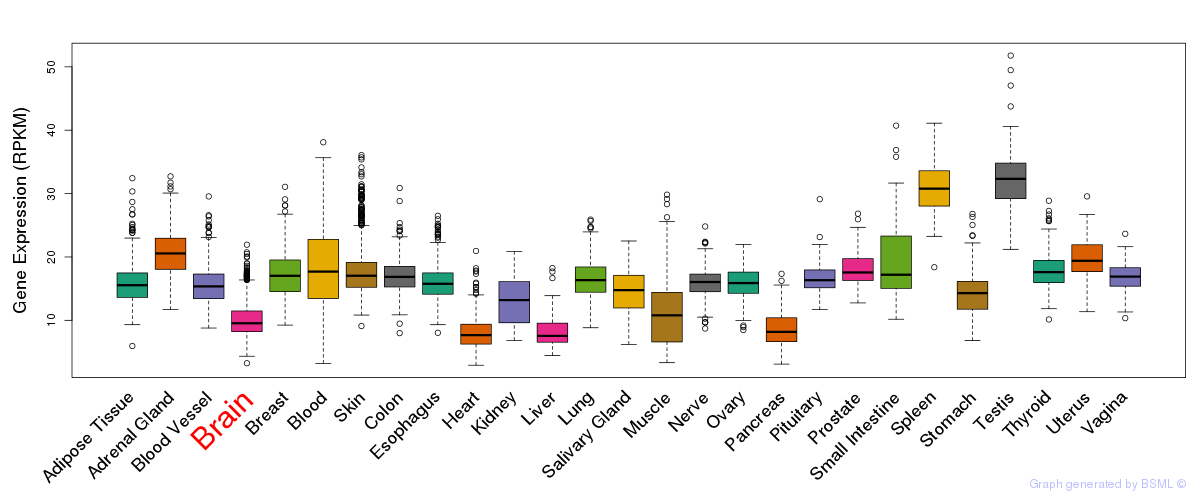
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ASPHD1 | 0.81 | 0.85 |
| CCDC85B | 0.76 | 0.86 |
| RPS6KA4 | 0.75 | 0.79 |
| GNPTG | 0.75 | 0.82 |
| CYP46A1 | 0.75 | 0.84 |
| ALKBH6 | 0.74 | 0.79 |
| PGP | 0.73 | 0.74 |
| CEBPB | 0.72 | 0.78 |
| C17orf89 | 0.72 | 0.79 |
| NRGN | 0.72 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.61 | -0.57 |
| TUBB2B | -0.61 | -0.59 |
| CCDC123 | -0.60 | -0.58 |
| ZNF311 | -0.60 | -0.61 |
| THOC2 | -0.60 | -0.68 |
| ZNF551 | -0.60 | -0.60 |
| FNBP1L | -0.60 | -0.58 |
| AC004017.1 | -0.59 | -0.60 |
| TTC27 | -0.59 | -0.60 |
| KCNRG | -0.59 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005057 | receptor signaling protein activity | TAS | 9215629 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0016564 | transcription repressor activity | IEA | - | |
| GO:0031072 | heat shock protein binding | TAS | 15572661 | |
| GO:0042802 | identical protein binding | IPI | 10698492 | |
| GO:0047485 | protein N-terminus binding | IPI | 15572661 | |
| GO:0050681 | androgen receptor binding | IPI | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000281 | cytokinesis after mitosis | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008625 | induction of apoptosis via death domain receptors | TAS | 9215629 | |
| GO:0007257 | activation of JNK activity | TAS | 9215629 | |
| GO:0006915 | apoptosis | TAS | 15572661 | |
| GO:0030521 | androgen receptor signaling pathway | IDA | 15572661 | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 15572661 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000792 | heterochromatin | IEA | - | |
| GO:0005634 | nucleus | IDA | 15572661 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 15572661 | |
| GO:0016605 | PML body | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15572661 |
| CENPC1 | CENP-C | CENPC | MIF2 | hcp-4 | centromere protein C 1 | - | HPRD,BioGRID | 9645950 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | c-FLIP(L) interacts with Daxx. | BIND | 14637155 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with Daxx. | BIND | 11799127 |
| DAPK3 | FLJ36473 | ZIP | ZIPK | death-associated protein kinase 3 | - | HPRD,BioGRID | 12917339 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD,BioGRID | 10698492 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | Daxx interacts with p51-ETS1. | BIND | 10698492 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | Daxx interacts with p42-ETS1. | BIND | 10698492 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Reconstituted Complex | BioGRID | 11483955 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 9215629 |11193028 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD | 9215629 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 10669754 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Far Western | BioGRID | 10669754 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Far Western | BioGRID | 10669754 |
| HIPK1 | KIAA0630 | MGC26642 | MGC33446 | MGC33548 | Myak | Nbak2 | homeodomain interacting protein kinase 1 | - | HPRD,BioGRID | 12529400 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD,BioGRID | 14678985 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 9743501 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | - | HPRD,BioGRID | 11948183 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | Daxx interacts with MCRS1 (MSP58). | BIND | 11948183 |
| MX1 | IFI-78K | IFI78 | MX | MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | in vitro Two-hybrid | BioGRID | 11716541 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12595526 |
| PAX3 | CDHS | HUP2 | MGC120381 | MGC120382 | MGC120383 | MGC120384 | MGC134778 | WS1 | paired box 3 | - | HPRD,BioGRID | 10393185 |
| PAX5 | BSAP | paired box 5 | - | HPRD,BioGRID | 11799127 |
| PAX5 | BSAP | paired box 5 | Pax5 interacts with Daxx. | BIND | 11799127 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 10525530 |10669754 |10684855 |12595526 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Affinity Capture-Western Co-localization Two-hybrid | BioGRID | 10525530 |10669754 |10684855 |11244500 |
| SLC2A4 | GLUT4 | solute carrier family 2 (facilitated glucose transporter), member 4 | - | HPRD,BioGRID | 11842083 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Daxx interacts with sentrin. | BIND | 11112409 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD,BioGRID | 11112409 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Daxx interacts with an unspecified isoform of SUMO1. | BIND | 11948183 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD | 11483955 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | Reconstituted Complex | BioGRID | 11483955 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD | 11483955 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Daxx interacts with Ubc9. | BIND | 11112409 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Biochemical Activity Two-hybrid | BioGRID | 11112409 |18691969 |