Gene Page: DCN
Summary ?
| GeneID | 1634 |
| Symbol | DCN |
| Synonyms | CSCD|DSPG2|PG40|PGII|PGS2|SLRR1B |
| Description | decorin |
| Reference | MIM:125255|HGNC:HGNC:2705|Ensembl:ENSG00000011465|HPRD:00501| |
| Gene type | protein-coding |
| Map location | 12q21.33 |
| Pascal p-value | 0.08 |
| Sherlock p-value | 0.01 |
| Fetal beta | -0.769 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0119 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16825495 | chr2 | 134234401 | DCN | 1634 | 0.18 | trans | ||
| rs2922180 | chr3 | 125280472 | DCN | 1634 | 0.02 | trans | ||
| rs2976809 | chr3 | 125451738 | DCN | 1634 | 0.04 | trans | ||
| rs16894149 | chr5 | 61013724 | DCN | 1634 | 0.03 | trans | ||
| rs816736 | chr5 | 154271947 | DCN | 1634 | 0.04 | trans | ||
| rs6934246 | chr6 | 35154494 | DCN | 1634 | 0.1 | trans | ||
| rs4723002 | chr7 | 30725700 | DCN | 1634 | 0.08 | trans | ||
| rs4723003 | chr7 | 30725740 | DCN | 1634 | 0.08 | trans | ||
| rs2199402 | chr8 | 9201002 | DCN | 1634 | 0.04 | trans | ||
| rs7841407 | chr8 | 9243427 | DCN | 1634 | 0.01 | trans | ||
| rs17134872 | chr11 | 76139845 | DCN | 1634 | 0.19 | trans | ||
| rs11236774 | chr11 | 76234952 | DCN | 1634 | 0.19 | trans | ||
| rs17720221 | chr13 | 75182377 | DCN | 1634 | 0.19 | trans | ||
| rs10149864 | chr14 | 86719683 | DCN | 1634 | 0.08 | trans | ||
| rs2253733 | chr15 | 66299343 | DCN | 1634 | 0.17 | trans |
Section II. Transcriptome annotation
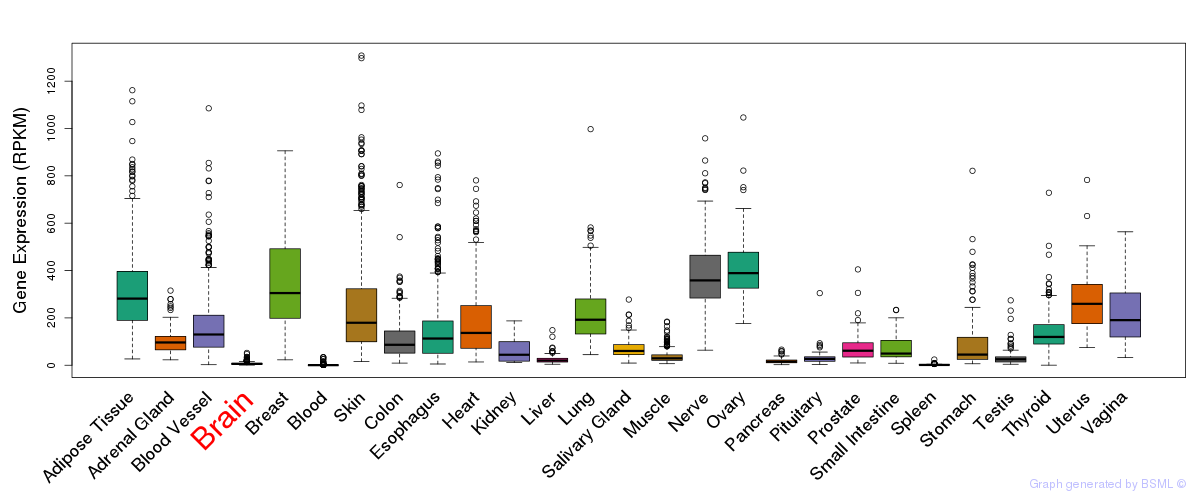
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AKR1C1 | 0.92 | 0.95 |
| STMN2 | 0.83 | 0.91 |
| ACAT2 | 0.82 | 0.90 |
| TUBA1A | 0.82 | 0.91 |
| TUBB3 | 0.82 | 0.91 |
| COTL1 | 0.82 | 0.84 |
| CDC42 | 0.81 | 0.90 |
| TSPAN2 | 0.81 | 0.87 |
| SMAP1 | 0.80 | 0.87 |
| PPP3CC | 0.80 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.66 | -0.75 |
| HEPN1 | -0.62 | -0.73 |
| SPARCL1 | -0.62 | -0.67 |
| AF347015.27 | -0.62 | -0.81 |
| AF347015.33 | -0.62 | -0.81 |
| SLC16A11 | -0.61 | -0.64 |
| AF347015.31 | -0.61 | -0.77 |
| MT-CO2 | -0.60 | -0.78 |
| AF347015.8 | -0.59 | -0.80 |
| TSC22D4 | -0.59 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009887 | organ morphogenesis | TAS | 7961765 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHSG | A2HS | AHS | FETUA | HSGA | alpha-2-HS-glycoprotein | - | HPRD,BioGRID | 12071714 |
| C1QA | - | complement component 1, q subcomponent, A chain | - | HPRD,BioGRID | 1431141 |
| COL14A1 | UND | collagen, type XIV, alpha 1 | - | HPRD,BioGRID | 9252349 |
| COL1A1 | OI4 | collagen, type I, alpha 1 | - | HPRD | 1468447 |9675033 |
| COL1A2 | OI4 | collagen, type I, alpha 2 | - | HPRD,BioGRID | 1468447 |2375748 |9675033 |
| COL2A1 | ANFH | AOM | COL11A3 | MGC131516 | SEDC | collagen, type II, alpha 1 | - | HPRD | 10382266 |
| COL4A1 | arresten | collagen, type IV, alpha 1 | - | HPRD | 10382266 |
| COL4A2 | DKFZp686I14213 | FLJ22259 | collagen, type IV, alpha 2 | - | HPRD | 10382266 |
| COL4A3 | - | collagen, type IV, alpha 3 (Goodpasture antigen) | - | HPRD | 10382266 |
| COL4A4 | CA44 | collagen, type IV, alpha 4 | - | HPRD | 10382266 |
| COL4A5 | ASLN | ATS | CA54 | MGC167109 | MGC42377 | collagen, type IV, alpha 5 | - | HPRD | 10382266 |
| COL4A6 | MGC88184 | collagen, type IV, alpha 6 | - | HPRD | 10382266 |
| COL6A1 | OPLL | collagen, type VI, alpha 1 | Reconstituted Complex | BioGRID | 1544908 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9988678 |12105206 |
| ELN | FLJ38671 | FLJ43523 | SVAS | WBS | WS | elastin | - | HPRD,BioGRID | 11723132 |
| FBN1 | FBN | MASS | MFS1 | OCTD | SGS | WMS | fibrillin 1 | - | HPRD | 10793130 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 12106908 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD | 1468447 |
| PLA2G2A | MOM1 | PLA2 | PLA2B | PLA2L | PLA2S | PLAS1 | sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) | - | HPRD,BioGRID | 10747008 |
| SFTPA1B | AC068139.6 | MGC133365 | PSAP | PSPA | SFTP1 | SFTPA1 | surfactant protein A1B | Reconstituted Complex | BioGRID | 12730206 |
| SFTPD | COLEC7 | PSP-D | SFTP4 | SP-D | surfactant protein D | - | HPRD,BioGRID | 12730206 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD | 7638106 |7798269 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | Reconstituted Complex | BioGRID | 7798269 |8093006 |9675033 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | Reconstituted Complex | BioGRID | 9675033 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD | 1550960 |9328841 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | - | HPRD,BioGRID | 12387878 |
| WISP1 | CCN4 | WISP1c | WISP1i | WISP1tc | WNT1 inducible signaling pathway protein 1 | - | HPRD | 11598131 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME CS DS DEGRADATION | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR MARKERS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST NORMAL DUCTAL VS LOBULAR DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| NADERI BREAST CANCER PROGNOSIS DN | 18 | 13 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA FOREVER UP | 19 | 13 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA | 39 | 25 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 40 MCF10A | 32 | 21 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 2 UP | 14 | 13 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS DN | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN | 108 | 64 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL DN | 29 | 11 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP | 76 | 46 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| ZHU SKIL TARGETS UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-376c | 468 | 474 | 1A | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-496 | 470 | 476 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.