Gene Page: DLX6
Summary ?
| GeneID | 1750 |
| Symbol | DLX6 |
| Synonyms | - |
| Description | distal-less homeobox 6 |
| Reference | MIM:600030|HGNC:HGNC:2919|Ensembl:ENSG00000006377|HPRD:02493|Vega:OTTHUMG00000154201 |
| Gene type | protein-coding |
| Map location | 7q22 |
| Pascal p-value | 0.102 |
| Fetal beta | -0.504 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03774463 | 7 | 96636616 | DLX6AS;DLX6 | 9.27E-7 | -0.398 | 0.007 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
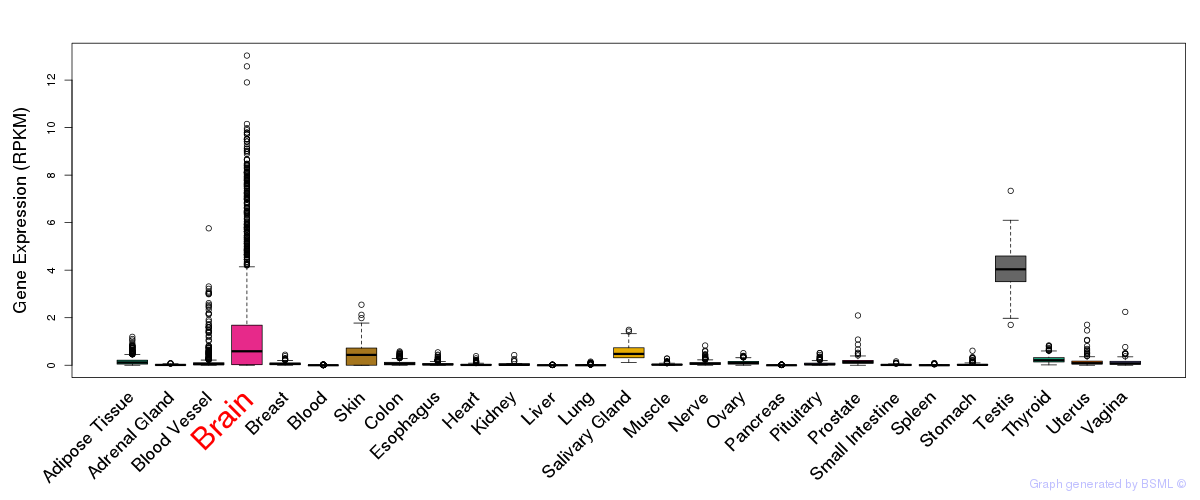
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| JPH4 | 0.90 | 0.90 |
| NUMBL | 0.89 | 0.89 |
| DUSP8 | 0.89 | 0.89 |
| MAPK8IP1 | 0.88 | 0.88 |
| MAPK8IP2 | 0.88 | 0.89 |
| SEMA6B | 0.88 | 0.89 |
| GIT1 | 0.88 | 0.87 |
| EPN1 | 0.88 | 0.88 |
| C19orf26 | 0.87 | 0.90 |
| STMN3 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.60 | -0.46 |
| MT-CO2 | -0.58 | -0.45 |
| AP002478.3 | -0.57 | -0.52 |
| AF347015.31 | -0.57 | -0.45 |
| NOSTRIN | -0.57 | -0.49 |
| AF347015.8 | -0.57 | -0.44 |
| AL139819.3 | -0.57 | -0.54 |
| AF347015.33 | -0.55 | -0.42 |
| MT-CYB | -0.55 | -0.43 |
| AF347015.2 | -0.55 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 7907794 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 7907794 |
| GO:0001501 | skeletal system development | TAS | 7907794 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030326 | embryonic limb morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7Q21 Q22 AMPLICON | 76 | 33 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-145 | 784 | 791 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-203.1 | 900 | 906 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-33 | 701 | 707 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-330 | 832 | 839 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-505 | 916 | 922 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.