Gene Page: DYNC1I2
Summary ?
| GeneID | 1781 |
| Symbol | DYNC1I2 |
| Synonyms | DIC74|DNCI2|IC2 |
| Description | dynein cytoplasmic 1 intermediate chain 2 |
| Reference | MIM:603331|HGNC:HGNC:2964|HPRD:04512| |
| Gene type | protein-coding |
| Map location | 2q31.1 |
| Pascal p-value | 1.327E-5 |
| Sherlock p-value | 0.048 |
| Fetal beta | 2.121 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1076773 | chr1 | 37136422 | DYNC1I2 | 1781 | 0.16 | trans | ||
| rs6829546 | chr4 | 12587664 | DYNC1I2 | 1781 | 0.07 | trans | ||
| rs6448932 | chr4 | 12595798 | DYNC1I2 | 1781 | 0.14 | trans |
Section II. Transcriptome annotation
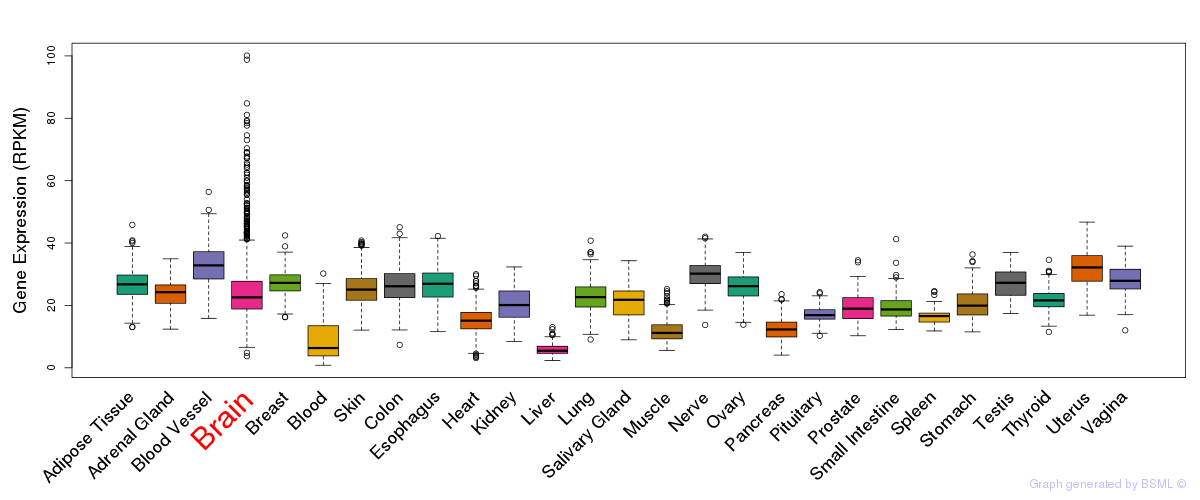
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SSFA2 | 0.87 | 0.87 |
| TLN1 | 0.87 | 0.86 |
| ITPKB | 0.86 | 0.87 |
| KANK1 | 0.86 | 0.84 |
| TJP1 | 0.86 | 0.82 |
| FGFR2 | 0.85 | 0.84 |
| OTUD7B | 0.84 | 0.82 |
| SASH1 | 0.84 | 0.82 |
| MTMR10 | 0.84 | 0.82 |
| ZCCHC24 | 0.84 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.49 | -0.53 |
| ZNF32 | -0.47 | -0.54 |
| COMMD3 | -0.47 | -0.53 |
| NR2C2AP | -0.47 | -0.51 |
| AC011491.1 | -0.47 | -0.49 |
| BID | -0.47 | -0.50 |
| DCTN3 | -0.47 | -0.48 |
| NDUFAF2 | -0.46 | -0.53 |
| POLB | -0.46 | -0.51 |
| C3orf54 | -0.45 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |