Gene Page: DOCK3
Summary ?
| GeneID | 1795 |
| Symbol | DOCK3 |
| Synonyms | MOCA|PBP |
| Description | dedicator of cytokinesis 3 |
| Reference | MIM:603123|HGNC:HGNC:2989|Ensembl:ENSG00000088538|HPRD:10341|Vega:OTTHUMG00000156892 |
| Gene type | protein-coding |
| Map location | 3p21.2 |
| Pascal p-value | 0.302 |
| Fetal beta | -0.527 |
| eGene | Nucleus accumbens basal ganglia |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
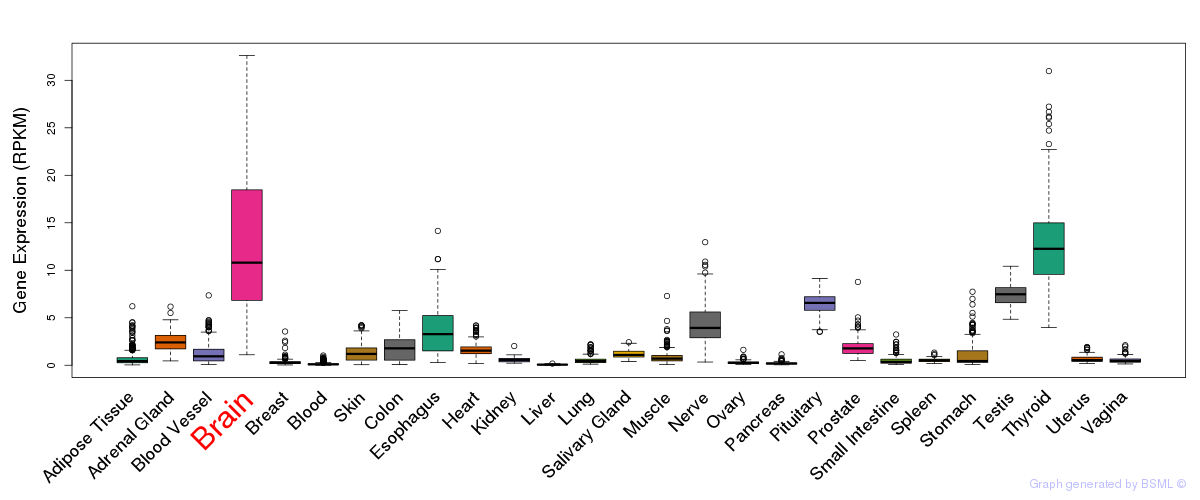
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMPX | 0.83 | 0.60 |
| CASQ2 | 0.82 | 0.45 |
| ABHD12B | 0.81 | 0.71 |
| WNT2 | 0.81 | 0.57 |
| CARD11 | 0.79 | 0.63 |
| RDH12 | 0.79 | 0.72 |
| TTC39A | 0.78 | 0.75 |
| FAM19A4 | 0.78 | 0.68 |
| MCF2 | 0.77 | 0.65 |
| NEXN | 0.77 | 0.33 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR86 | -0.35 | -0.42 |
| CARHSP1 | -0.31 | -0.47 |
| DACT1 | -0.31 | -0.26 |
| SLA | -0.30 | -0.24 |
| EIF4EBP1 | -0.30 | -0.45 |
| MYCN | -0.30 | -0.26 |
| TTC28 | -0.30 | -0.33 |
| KIAA1949 | -0.30 | -0.39 |
| ZNF238 | -0.30 | -0.27 |
| NEUROD2 | -0.30 | -0.35 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR UP | 25 | 17 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |