Gene Page: EGR2
Summary ?
| GeneID | 1959 |
| Symbol | EGR2 |
| Synonyms | AT591|CMT1D|CMT4E|KROX20 |
| Description | early growth response 2 |
| Reference | MIM:129010|HGNC:HGNC:3239|Ensembl:ENSG00000122877|HPRD:00551|Vega:OTTHUMG00000018308 |
| Gene type | protein-coding |
| Map location | 10q21.1 |
| Pascal p-value | 0.061 |
| Fetal beta | -1.595 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16918799 | chr10 | 65464025 | EGR2 | 1959 | 0.14 | cis | ||
| rs307378 | 0 | EGR2 | 1959 | 0.19 | trans | |||
| rs6677917 | chr1 | 34406841 | EGR2 | 1959 | 0.14 | trans | ||
| rs983619 | chr1 | 39047418 | EGR2 | 1959 | 0.05 | trans | ||
| rs7545984 | chr1 | 45003892 | EGR2 | 1959 | 0.01 | trans | ||
| rs7554123 | chr1 | 45004203 | EGR2 | 1959 | 0.01 | trans | ||
| rs12059674 | chr1 | 57630277 | EGR2 | 1959 | 0.02 | trans | ||
| rs17126801 | chr1 | 65098162 | EGR2 | 1959 | 0.02 | trans | ||
| rs17474410 | chr1 | 68267242 | EGR2 | 1959 | 0.14 | trans | ||
| rs7511937 | 0 | EGR2 | 1959 | 0 | trans | |||
| rs2782784 | chr1 | 71195282 | EGR2 | 1959 | 0.01 | trans | ||
| rs2257700 | chr1 | 71273516 | EGR2 | 1959 | 0.05 | trans | ||
| rs7541092 | chr1 | 71362471 | EGR2 | 1959 | 0.09 | trans | ||
| rs17689089 | chr1 | 77740105 | EGR2 | 1959 | 0 | trans | ||
| rs17100231 | chr1 | 77761339 | EGR2 | 1959 | 0.01 | trans | ||
| rs17100274 | chr1 | 77774412 | EGR2 | 1959 | 3.806E-5 | trans | ||
| rs12034005 | chr1 | 79793034 | EGR2 | 1959 | 0.04 | trans | ||
| rs3015033 | chr1 | 79847235 | EGR2 | 1959 | 0.01 | trans | ||
| rs17098656 | chr1 | 80098386 | EGR2 | 1959 | 0.01 | trans | ||
| rs6664544 | chr1 | 92524192 | EGR2 | 1959 | 0 | trans | ||
| rs12403481 | chr1 | 109503655 | EGR2 | 1959 | 0 | trans | ||
| rs337294 | 0 | EGR2 | 1959 | 0.02 | trans | |||
| rs17029293 | chr1 | 112567844 | EGR2 | 1959 | 2.975E-4 | trans | ||
| rs16837984 | chr1 | 157042626 | EGR2 | 1959 | 0 | trans | ||
| rs16837989 | chr1 | 157046286 | EGR2 | 1959 | 0.01 | trans | ||
| rs16838012 | chr1 | 157048974 | EGR2 | 1959 | 0 | trans | ||
| rs16838014 | 0 | EGR2 | 1959 | 0 | trans | |||
| rs4657385 | chr1 | 164939279 | EGR2 | 1959 | 0.14 | trans | ||
| rs6666937 | chr1 | 170286842 | EGR2 | 1959 | 8.539E-4 | trans | ||
| rs12071422 | chr1 | 185573879 | EGR2 | 1959 | 0.01 | trans | ||
| rs16843112 | chr1 | 198420657 | EGR2 | 1959 | 0.19 | trans | ||
| rs697455 | chr1 | 202512075 | EGR2 | 1959 | 0.02 | trans | ||
| rs4457633 | chr1 | 204283819 | EGR2 | 1959 | 0.04 | trans | ||
| rs3748669 | chr1 | 207077022 | EGR2 | 1959 | 0.01 | trans | ||
| rs6673109 | chr1 | 207253132 | EGR2 | 1959 | 9.237E-5 | trans | ||
| rs17260245 | chr1 | 209411024 | EGR2 | 1959 | 0.17 | trans | ||
| rs1796829 | chr1 | 239661818 | EGR2 | 1959 | 3.155E-4 | trans | ||
| rs12034521 | chr1 | 239717679 | EGR2 | 1959 | 1.996E-5 | trans | ||
| rs6670526 | chr1 | 239725414 | EGR2 | 1959 | 0.16 | trans | ||
| snp_a-1990354 | 0 | EGR2 | 1959 | 9.225E-7 | trans | |||
| rs939217 | chr2 | 12730751 | EGR2 | 1959 | 0.03 | trans | ||
| rs6432434 | chr2 | 14160930 | EGR2 | 1959 | 0 | trans | ||
| rs6712107 | chr2 | 14178653 | EGR2 | 1959 | 0.03 | trans | ||
| rs16861129 | chr2 | 14201738 | EGR2 | 1959 | 0.03 | trans | ||
| rs16861131 | chr2 | 14202085 | EGR2 | 1959 | 0.03 | trans | ||
| rs7589712 | chr2 | 14205064 | EGR2 | 1959 | 0.14 | trans | ||
| rs12329305 | chr2 | 19552912 | EGR2 | 1959 | 0 | trans | ||
| rs12611832 | chr2 | 31196248 | EGR2 | 1959 | 0.02 | trans | ||
| rs12614732 | chr2 | 31199913 | EGR2 | 1959 | 0.02 | trans | ||
| rs17023938 | chr2 | 39773236 | EGR2 | 1959 | 0.18 | trans | ||
| rs7560081 | chr2 | 62793238 | EGR2 | 1959 | 0 | trans | ||
| rs17029079 | chr2 | 64736071 | EGR2 | 1959 | 0.01 | trans | ||
| rs17038084 | chr2 | 85192280 | EGR2 | 1959 | 4.202E-5 | trans | ||
| rs17024035 | chr2 | 100968559 | EGR2 | 1959 | 0.06 | trans | ||
| rs7574684 | chr2 | 100992911 | EGR2 | 1959 | 0.06 | trans | ||
| rs17024129 | chr2 | 100995144 | EGR2 | 1959 | 0.06 | trans | ||
| rs7585898 | chr2 | 100998292 | EGR2 | 1959 | 0.06 | trans | ||
| rs1370627 | chr2 | 101009384 | EGR2 | 1959 | 0.06 | trans | ||
| rs17042517 | chr2 | 113639583 | EGR2 | 1959 | 0.18 | trans | ||
| rs17047906 | chr2 | 119007824 | EGR2 | 1959 | 0.12 | trans | ||
| rs1840209 | chr2 | 125625401 | EGR2 | 1959 | 0.02 | trans | ||
| rs6707117 | chr2 | 136855504 | EGR2 | 1959 | 2.06E-12 | trans | ||
| rs17834293 | chr2 | 142993878 | EGR2 | 1959 | 0.14 | trans | ||
| rs7564795 | chr2 | 143033291 | EGR2 | 1959 | 0.09 | trans | ||
| rs10189091 | chr2 | 143471999 | EGR2 | 1959 | 9.547E-4 | trans | ||
| snp_a-2198363 | 0 | EGR2 | 1959 | 0.01 | trans | |||
| rs707097 | chr2 | 155318814 | EGR2 | 1959 | 0.03 | trans | ||
| rs16838911 | chr2 | 156087569 | EGR2 | 1959 | 3.899E-4 | trans | ||
| rs12469388 | chr2 | 178689030 | EGR2 | 1959 | 0.19 | trans | ||
| rs7608942 | chr2 | 182820100 | EGR2 | 1959 | 0.03 | trans | ||
| rs16848675 | chr2 | 213349996 | EGR2 | 1959 | 0.04 | trans | ||
| rs16829292 | chr2 | 233447622 | EGR2 | 1959 | 0.01 | trans | ||
| rs16829401 | chr2 | 233538391 | EGR2 | 1959 | 0 | trans | ||
| rs17040622 | chr3 | 4440673 | EGR2 | 1959 | 0.01 | trans | ||
| rs17684934 | chr3 | 4443547 | EGR2 | 1959 | 1.114E-4 | trans | ||
| rs17040633 | chr3 | 4446836 | EGR2 | 1959 | 1.114E-4 | trans | ||
| rs17756483 | chr3 | 4451309 | EGR2 | 1959 | 1.114E-4 | trans | ||
| rs17685441 | chr3 | 4465165 | EGR2 | 1959 | 1.114E-4 | trans | ||
| rs17015415 | chr3 | 24933888 | EGR2 | 1959 | 0.03 | trans | ||
| rs12639503 | chr3 | 51860816 | EGR2 | 1959 | 0.06 | trans | ||
| rs9863778 | chr3 | 55375950 | EGR2 | 1959 | 0.02 | trans | ||
| rs997245 | chr3 | 61844600 | EGR2 | 1959 | 8.765E-4 | trans | ||
| rs3796213 | chr3 | 69157380 | EGR2 | 1959 | 0.05 | trans | ||
| rs6549186 | chr3 | 69211954 | EGR2 | 1959 | 0.07 | trans | ||
| rs2875492 | chr3 | 86006209 | EGR2 | 1959 | 0.01 | trans | ||
| rs16843128 | chr3 | 100713413 | EGR2 | 1959 | 6.137E-4 | trans | ||
| rs1381835 | chr3 | 110600595 | EGR2 | 1959 | 0 | trans | ||
| rs1877965 | chr3 | 126013904 | EGR2 | 1959 | 0.15 | trans | ||
| rs9289277 | chr3 | 126038339 | EGR2 | 1959 | 0.1 | trans | ||
| rs16839796 | chr4 | 7188348 | EGR2 | 1959 | 7.519E-5 | trans | ||
| rs4631023 | chr4 | 8101794 | EGR2 | 1959 | 0.07 | trans | ||
| rs7684087 | chr4 | 21673380 | EGR2 | 1959 | 0.01 | trans | ||
| rs16989064 | chr4 | 33639397 | EGR2 | 1959 | 0 | trans | ||
| rs11096966 | chr4 | 38961258 | EGR2 | 1959 | 0.13 | trans | ||
| rs4469113 | chr4 | 41461062 | EGR2 | 1959 | 2.509E-4 | trans | ||
| rs6836259 | chr4 | 57173074 | EGR2 | 1959 | 0.06 | trans | ||
| rs2643038 | chr4 | 62313218 | EGR2 | 1959 | 0.01 | trans | ||
| snp_a-1830437 | 0 | EGR2 | 1959 | 0.15 | trans | |||
| rs6817626 | chr4 | 78005836 | EGR2 | 1959 | 0.04 | trans | ||
| rs1604153 | chr4 | 78472744 | EGR2 | 1959 | 0.02 | trans | ||
| rs1564462 | chr4 | 79910988 | EGR2 | 1959 | 0.08 | trans | ||
| rs6827713 | chr4 | 79936877 | EGR2 | 1959 | 0.02 | trans | ||
| rs17005000 | chr4 | 81826551 | EGR2 | 1959 | 0.04 | trans | ||
| rs10011751 | chr4 | 93984994 | EGR2 | 1959 | 0.01 | trans | ||
| rs11099888 | chr4 | 154498555 | EGR2 | 1959 | 0.07 | trans | ||
| rs17701328 | chr4 | 178744532 | EGR2 | 1959 | 0.17 | trans | ||
| rs6881821 | chr5 | 8578465 | EGR2 | 1959 | 8.795E-5 | trans | ||
| rs9312843 | chr5 | 13802997 | EGR2 | 1959 | 0 | trans | ||
| rs1732944 | chr5 | 30905595 | EGR2 | 1959 | 0.11 | trans | ||
| rs1697073 | chr5 | 30907267 | EGR2 | 1959 | 0.01 | trans | ||
| rs9292700 | chr5 | 38192610 | EGR2 | 1959 | 0.18 | trans | ||
| rs2115110 | chr5 | 38193428 | EGR2 | 1959 | 1.764E-5 | trans | ||
| rs16873956 | chr5 | 44312488 | EGR2 | 1959 | 0 | trans | ||
| rs10040715 | chr5 | 57594207 | EGR2 | 1959 | 5.105E-11 | trans | ||
| rs16894149 | chr5 | 61013724 | EGR2 | 1959 | 0.1 | trans | ||
| snp_a-4217300 | 0 | EGR2 | 1959 | 1.213E-6 | trans | |||
| rs1862599 | chr5 | 82095088 | EGR2 | 1959 | 0 | trans | ||
| rs163953 | chr5 | 92289536 | EGR2 | 1959 | 0.01 | trans | ||
| rs17134275 | chr5 | 100634998 | EGR2 | 1959 | 0 | trans | ||
| rs17257221 | chr5 | 101016609 | EGR2 | 1959 | 0.07 | trans | ||
| rs10063272 | chr5 | 103540974 | EGR2 | 1959 | 0.1 | trans | ||
| rs2963232 | chr5 | 103713882 | EGR2 | 1959 | 0.02 | trans | ||
| rs250323 | chr5 | 115646995 | EGR2 | 1959 | 0.03 | trans | ||
| rs1350578 | chr5 | 116310282 | EGR2 | 1959 | 0.02 | trans | ||
| rs2900115 | chr5 | 117414349 | EGR2 | 1959 | 0.11 | trans | ||
| rs11955842 | chr5 | 118066618 | EGR2 | 1959 | 0.04 | trans | ||
| rs6894229 | chr5 | 127160990 | EGR2 | 1959 | 0.18 | trans | ||
| rs2141482 | chr5 | 136760477 | EGR2 | 1959 | 8.057E-5 | trans | ||
| snp_a-1983671 | 0 | EGR2 | 1959 | 0.18 | trans | |||
| rs2189803 | chr5 | 137684661 | EGR2 | 1959 | 1.274E-8 | trans | ||
| rs17063216 | chr5 | 163777647 | EGR2 | 1959 | 0.02 | trans | ||
| rs17063224 | chr5 | 163780687 | EGR2 | 1959 | 0.02 | trans | ||
| rs17666252 | chr5 | 168294345 | EGR2 | 1959 | 0.19 | trans | ||
| snp_a-2055204 | 0 | EGR2 | 1959 | 0.04 | trans | |||
| rs259913 | chr5 | 169370782 | EGR2 | 1959 | 1.629E-5 | trans | ||
| snp_a-2275210 | 0 | EGR2 | 1959 | 0.17 | trans | |||
| rs2116811 | chr5 | 171820051 | EGR2 | 1959 | 3.122E-4 | trans | ||
| rs258875 | chr5 | 173046554 | EGR2 | 1959 | 0.05 | trans | ||
| rs16883859 | chr6 | 20474617 | EGR2 | 1959 | 0.09 | trans | ||
| rs16883862 | chr6 | 20474766 | EGR2 | 1959 | 0.09 | trans | ||
| rs10484635 | chr6 | 20538664 | EGR2 | 1959 | 0.05 | trans | ||
| rs7747254 | chr6 | 24013530 | EGR2 | 1959 | 0.14 | trans | ||
| rs3909134 | chr6 | 29699252 | EGR2 | 1959 | 0.13 | trans | ||
| rs195429 | chr6 | 37354434 | EGR2 | 1959 | 0.01 | trans | ||
| rs16890522 | chr6 | 38236301 | EGR2 | 1959 | 0.16 | trans | ||
| rs12528414 | chr6 | 38404800 | EGR2 | 1959 | 0.03 | trans | ||
| rs7764900 | chr6 | 38440495 | EGR2 | 1959 | 0.08 | trans | ||
| rs6915776 | chr6 | 71797961 | EGR2 | 1959 | 0.19 | trans | ||
| rs17069168 | chr6 | 108335432 | EGR2 | 1959 | 0.11 | trans | ||
| rs17069171 | chr6 | 108335593 | EGR2 | 1959 | 0.05 | trans | ||
| rs391196 | chr6 | 111563907 | EGR2 | 1959 | 0.06 | trans | ||
| rs4289676 | chr6 | 123999302 | EGR2 | 1959 | 7.47E-4 | trans | ||
| rs17055235 | chr6 | 128355263 | EGR2 | 1959 | 0.06 | trans | ||
| rs2030024 | chr6 | 131014624 | EGR2 | 1959 | 0 | trans | ||
| rs9402280 | chr6 | 131102770 | EGR2 | 1959 | 0.01 | trans | ||
| rs17062925 | chr6 | 134168614 | EGR2 | 1959 | 0.18 | trans | ||
| rs7767456 | chr6 | 134356119 | EGR2 | 1959 | 0.03 | trans | ||
| rs9385758 | chr6 | 136729593 | EGR2 | 1959 | 0.19 | trans | ||
| rs12176346 | chr6 | 136732663 | EGR2 | 1959 | 0 | trans | ||
| rs9402819 | chr6 | 136848169 | EGR2 | 1959 | 0.02 | trans | ||
| rs9389400 | chr6 | 136854803 | EGR2 | 1959 | 0.02 | trans | ||
| rs2237264 | chr6 | 136919977 | EGR2 | 1959 | 0 | trans | ||
| rs17080121 | chr6 | 151009189 | EGR2 | 1959 | 0 | trans | ||
| rs12529269 | chr6 | 153543829 | EGR2 | 1959 | 0 | trans | ||
| rs10872699 | chr6 | 153544252 | EGR2 | 1959 | 4.044E-5 | trans | ||
| rs1321606 | chr6 | 153545278 | EGR2 | 1959 | 0 | trans | ||
| rs10945911 | chr6 | 164047509 | EGR2 | 1959 | 1.075E-4 | trans | ||
| rs17156078 | chr7 | 27988688 | EGR2 | 1959 | 0 | trans | ||
| rs12701006 | chr7 | 30147298 | EGR2 | 1959 | 1.07E-6 | trans | ||
| rs1156490 | chr7 | 34334956 | EGR2 | 1959 | 0.17 | trans | ||
| rs1419793 | chr7 | 34697630 | EGR2 | 1959 | 0.19 | trans | ||
| rs17815964 | chr7 | 52540908 | EGR2 | 1959 | 0.04 | trans | ||
| rs17621151 | chr7 | 53572911 | EGR2 | 1959 | 1.204E-4 | trans | ||
| rs10245449 | chr7 | 80935100 | EGR2 | 1959 | 0.01 | trans | ||
| rs2272618 | chr7 | 107871360 | EGR2 | 1959 | 0.13 | trans | ||
| rs12669568 | chr7 | 107885942 | EGR2 | 1959 | 0.13 | trans | ||
| rs7777322 | chr7 | 107886663 | EGR2 | 1959 | 0.13 | trans | ||
| rs12671672 | chr7 | 117614219 | EGR2 | 1959 | 0.03 | trans | ||
| rs10487383 | chr7 | 117671737 | EGR2 | 1959 | 0 | trans | ||
| rs6958821 | chr7 | 153253566 | EGR2 | 1959 | 0 | trans | ||
| rs1865462 | chr7 | 157637961 | EGR2 | 1959 | 0.12 | trans | ||
| rs11779237 | chr8 | 1227664 | EGR2 | 1959 | 0.01 | trans | ||
| rs1358128 | chr8 | 5268072 | EGR2 | 1959 | 0.01 | trans | ||
| rs12682256 | chr8 | 5306138 | EGR2 | 1959 | 0.01 | trans | ||
| rs1010895 | chr8 | 14580230 | EGR2 | 1959 | 0.02 | trans | ||
| rs10109456 | chr8 | 19565941 | EGR2 | 1959 | 0.01 | trans | ||
| rs17054681 | chr8 | 25814323 | EGR2 | 1959 | 0.07 | trans | ||
| rs16877914 | chr8 | 31196384 | EGR2 | 1959 | 0.08 | trans | ||
| rs1457044 | chr8 | 59257567 | EGR2 | 1959 | 4.062E-7 | trans | ||
| rs1457032 | chr8 | 59290536 | EGR2 | 1959 | 4.955E-5 | trans | ||
| rs2680903 | chr8 | 59512333 | EGR2 | 1959 | 0.04 | trans | ||
| rs2634533 | chr8 | 59535621 | EGR2 | 1959 | 0.06 | trans | ||
| rs7465259 | 0 | EGR2 | 1959 | 0.01 | trans | |||
| rs10104622 | chr8 | 63333773 | EGR2 | 1959 | 0.01 | trans | ||
| rs2218911 | chr8 | 63371015 | EGR2 | 1959 | 0.01 | trans | ||
| rs1551199 | chr8 | 63388117 | EGR2 | 1959 | 0.01 | trans | ||
| rs1733950 | chr8 | 90062516 | EGR2 | 1959 | 2.51E-10 | trans | ||
| rs1240028 | chr8 | 90093357 | EGR2 | 1959 | 1.261E-4 | trans | ||
| rs1240087 | chr8 | 90113945 | EGR2 | 1959 | 5.906E-4 | trans | ||
| rs318307 | chr8 | 90185459 | EGR2 | 1959 | 1.775E-4 | trans | ||
| rs1483373 | chr8 | 90186442 | EGR2 | 1959 | 0.02 | trans | ||
| rs11776005 | chr8 | 90187416 | EGR2 | 1959 | 0 | trans | ||
| rs1159499 | chr8 | 90196948 | EGR2 | 1959 | 0.01 | trans | ||
| rs1355060 | chr8 | 118717710 | EGR2 | 1959 | 0 | trans | ||
| rs2280828 | chr8 | 118762435 | EGR2 | 1959 | 0 | trans | ||
| rs17683401 | chr8 | 119671834 | EGR2 | 1959 | 0.16 | trans | ||
| rs16901935 | chr8 | 128076544 | EGR2 | 1959 | 0.07 | trans | ||
| rs4976941 | chr8 | 142744439 | EGR2 | 1959 | 3.33E-5 | trans | ||
| rs9298651 | chr9 | 10076275 | EGR2 | 1959 | 1.259E-5 | trans | ||
| rs16931013 | chr9 | 10076859 | EGR2 | 1959 | 1.259E-5 | trans | ||
| rs1467547 | chr9 | 17630726 | EGR2 | 1959 | 0.01 | trans | ||
| rs4415417 | chr9 | 17837857 | EGR2 | 1959 | 0.06 | trans | ||
| rs2210327 | chr9 | 18109234 | EGR2 | 1959 | 0.03 | trans | ||
| rs6476383 | chr9 | 32865568 | EGR2 | 1959 | 0.02 | trans | ||
| rs11143622 | chr9 | 71370050 | EGR2 | 1959 | 0.01 | trans | ||
| rs11143868 | chr9 | 71437411 | EGR2 | 1959 | 0.06 | trans | ||
| rs11143995 | chr9 | 71462852 | EGR2 | 1959 | 0.06 | trans | ||
| rs11144031 | chr9 | 71477107 | EGR2 | 1959 | 0.06 | trans | ||
| rs11791891 | chr9 | 87512411 | EGR2 | 1959 | 0.01 | trans | ||
| rs2165893 | chr9 | 87531423 | EGR2 | 1959 | 0 | trans | ||
| rs763623 | chr9 | 87539663 | EGR2 | 1959 | 0.12 | trans | ||
| rs3780642 | chr9 | 87553144 | EGR2 | 1959 | 6.457E-6 | trans | ||
| rs10992752 | chr9 | 96240578 | EGR2 | 1959 | 0.01 | trans | ||
| rs10981864 | chr9 | 116416735 | EGR2 | 1959 | 0.1 | trans | ||
| snp_a-1879318 | 0 | EGR2 | 1959 | 0.09 | trans | |||
| rs2807706 | chr9 | 121744331 | EGR2 | 1959 | 2.853E-4 | trans | ||
| rs2818297 | chr9 | 121757015 | EGR2 | 1959 | 0.01 | trans | ||
| rs10739527 | chr9 | 121772512 | EGR2 | 1959 | 0.01 | trans | ||
| rs6537944 | chr9 | 137312827 | EGR2 | 1959 | 0.02 | trans | ||
| rs11103132 | chr9 | 138575901 | EGR2 | 1959 | 0.01 | trans | ||
| rs1017361 | chr10 | 294952 | EGR2 | 1959 | 0.16 | trans | ||
| rs6560840 | chr10 | 489655 | EGR2 | 1959 | 0.03 | trans | ||
| rs11252514 | chr10 | 499572 | EGR2 | 1959 | 8.532E-4 | trans | ||
| rs2924269 | chr10 | 4884783 | EGR2 | 1959 | 0.13 | trans | ||
| rs11014370 | chr10 | 25352798 | EGR2 | 1959 | 0.02 | trans | ||
| rs772394 | chr10 | 35980024 | EGR2 | 1959 | 0 | trans | ||
| rs17432714 | chr10 | 43526022 | EGR2 | 1959 | 0.05 | trans | ||
| rs2853836 | chr10 | 48431948 | EGR2 | 1959 | 0.02 | trans | ||
| rs1908351 | chr10 | 57877117 | EGR2 | 1959 | 0 | trans | ||
| rs3104871 | chr10 | 57878502 | EGR2 | 1959 | 7.348E-5 | trans | ||
| rs901309 | chr10 | 72571899 | EGR2 | 1959 | 0.01 | trans | ||
| rs4750827 | chr10 | 132547183 | EGR2 | 1959 | 0.03 | trans | ||
| rs4750829 | chr10 | 132547260 | EGR2 | 1959 | 0.04 | trans | ||
| rs11021918 | chr11 | 2269189 | EGR2 | 1959 | 0.01 | trans | ||
| rs862785 | chr11 | 20175604 | EGR2 | 1959 | 0.02 | trans | ||
| rs16911034 | chr11 | 23302320 | EGR2 | 1959 | 0 | trans | ||
| rs10838551 | chr11 | 46073112 | EGR2 | 1959 | 0.09 | trans | ||
| rs10838567 | chr11 | 46136342 | EGR2 | 1959 | 0.09 | trans | ||
| rs12292520 | chr11 | 48086150 | EGR2 | 1959 | 0.07 | trans | ||
| rs17142688 | chr11 | 81319064 | EGR2 | 1959 | 0.07 | trans | ||
| rs619320 | chr11 | 81846219 | EGR2 | 1959 | 0.13 | trans | ||
| rs11020043 | chr11 | 92470587 | EGR2 | 1959 | 0.06 | trans | ||
| rs2460047 | chr11 | 93678935 | EGR2 | 1959 | 0.18 | trans | ||
| rs1789300 | chr11 | 95463631 | EGR2 | 1959 | 4.92E-4 | trans | ||
| rs1727152 | chr11 | 95467435 | EGR2 | 1959 | 0.02 | trans | ||
| rs1255149 | chr11 | 95484493 | EGR2 | 1959 | 4.64E-4 | trans | ||
| rs3748251 | chr11 | 95519156 | EGR2 | 1959 | 1.302E-4 | trans | ||
| rs16922543 | chr11 | 95526116 | EGR2 | 1959 | 1.302E-4 | trans | ||
| rs16922622 | chr11 | 95567387 | EGR2 | 1959 | 1.302E-4 | trans | ||
| rs3181323 | chr11 | 104878386 | EGR2 | 1959 | 0.01 | trans | ||
| rs11219092 | chr11 | 123130750 | EGR2 | 1959 | 0.05 | trans | ||
| rs7119608 | chr11 | 134922824 | EGR2 | 1959 | 0.08 | trans | ||
| rs766095 | chr12 | 1498432 | EGR2 | 1959 | 0.15 | trans | ||
| rs2109556 | chr12 | 3887300 | EGR2 | 1959 | 0.14 | trans | ||
| rs733593 | chr12 | 6636391 | EGR2 | 1959 | 0.01 | trans | ||
| rs2058798 | chr12 | 16743164 | EGR2 | 1959 | 0.12 | trans | ||
| rs374484 | chr12 | 27627159 | EGR2 | 1959 | 8.488E-4 | trans | ||
| rs451921 | chr12 | 27627499 | EGR2 | 1959 | 0.11 | trans | ||
| rs10771436 | chr12 | 28744556 | EGR2 | 1959 | 0.06 | trans | ||
| rs1344796 | chr12 | 76397104 | EGR2 | 1959 | 3.074E-4 | trans | ||
| rs11831213 | chr12 | 109895355 | EGR2 | 1959 | 0.11 | trans | ||
| rs10850201 | chr12 | 109903061 | EGR2 | 1959 | 0 | trans | ||
| rs7133714 | chr12 | 120141546 | EGR2 | 1959 | 0.08 | trans | ||
| rs11833075 | chr12 | 122114592 | EGR2 | 1959 | 9.326E-5 | trans | ||
| rs1041050 | chr13 | 26585121 | EGR2 | 1959 | 0.01 | trans | ||
| rs9581602 | chr13 | 26834548 | EGR2 | 1959 | 0.03 | trans | ||
| rs17084271 | chr13 | 27089799 | EGR2 | 1959 | 0.03 | trans | ||
| rs17084280 | chr13 | 27090872 | EGR2 | 1959 | 0.03 | trans | ||
| rs9569893 | chr13 | 34198563 | EGR2 | 1959 | 0.09 | trans | ||
| rs2408301 | chr13 | 51808915 | EGR2 | 1959 | 0.08 | trans | ||
| rs7982110 | chr13 | 59723428 | EGR2 | 1959 | 6.427E-4 | trans | ||
| rs9528250 | chr13 | 61410190 | EGR2 | 1959 | 0 | trans | ||
| rs7330331 | chr13 | 67403122 | EGR2 | 1959 | 5.763E-4 | trans | ||
| rs17088392 | chr13 | 72257235 | EGR2 | 1959 | 0.04 | trans | ||
| rs7327697 | chr13 | 83227456 | EGR2 | 1959 | 7.968E-5 | trans | ||
| rs17667894 | chr13 | 92014308 | EGR2 | 1959 | 0 | trans | ||
| rs223110 | chr14 | 23973042 | EGR2 | 1959 | 0.08 | trans | ||
| rs7156088 | chr14 | 27821865 | EGR2 | 1959 | 6.427E-4 | trans | ||
| rs8013705 | chr14 | 34853284 | EGR2 | 1959 | 0.08 | trans | ||
| rs17106703 | chr14 | 37851979 | EGR2 | 1959 | 0.16 | trans | ||
| rs7147321 | chr14 | 45877904 | EGR2 | 1959 | 0.01 | trans | ||
| rs1952728 | chr14 | 45893091 | EGR2 | 1959 | 0.01 | trans | ||
| rs17116137 | chr14 | 45940865 | EGR2 | 1959 | 0.01 | trans | ||
| rs7151829 | chr14 | 45971281 | EGR2 | 1959 | 0.01 | trans | ||
| rs8022005 | chr14 | 61345380 | EGR2 | 1959 | 0 | trans | ||
| rs17100844 | chr14 | 63669261 | EGR2 | 1959 | 0.12 | trans | ||
| rs10148418 | chr14 | 67003872 | EGR2 | 1959 | 0 | trans | ||
| rs149642 | chr14 | 72653498 | EGR2 | 1959 | 0.02 | trans | ||
| rs3861624 | chr14 | 78416193 | EGR2 | 1959 | 0.07 | trans | ||
| rs941539 | chr14 | 93561214 | EGR2 | 1959 | 0.05 | trans | ||
| rs1885073 | chr14 | 104357181 | EGR2 | 1959 | 3.959E-6 | trans | ||
| rs4465541 | chr14 | 105016846 | EGR2 | 1959 | 0.09 | trans | ||
| rs16973929 | chr15 | 47395709 | EGR2 | 1959 | 0.01 | trans | ||
| rs8039808 | chr15 | 49939845 | EGR2 | 1959 | 0.09 | trans | ||
| rs10518936 | chr15 | 58090522 | EGR2 | 1959 | 0.04 | trans | ||
| rs2414703 | chr15 | 61904602 | EGR2 | 1959 | 0.01 | trans | ||
| rs289171 | chr15 | 62576053 | EGR2 | 1959 | 5.831E-4 | trans | ||
| rs289161 | chr15 | 62589888 | EGR2 | 1959 | 9.705E-4 | trans | ||
| rs939043 | chr15 | 70236428 | EGR2 | 1959 | 0.19 | trans | ||
| rs11858762 | chr15 | 81386594 | EGR2 | 1959 | 0.03 | trans | ||
| rs9635390 | chr15 | 90809632 | EGR2 | 1959 | 2.566E-5 | trans | ||
| rs7174562 | chr15 | 93811530 | EGR2 | 1959 | 0.07 | trans | ||
| rs16948359 | chr15 | 94233206 | EGR2 | 1959 | 0.19 | trans | ||
| rs16948375 | chr15 | 94236354 | EGR2 | 1959 | 0.03 | trans | ||
| rs17140355 | chr16 | 6548831 | EGR2 | 1959 | 0.12 | trans | ||
| rs8047151 | chr16 | 17209696 | EGR2 | 1959 | 1.871E-7 | trans | ||
| rs4271578 | chr16 | 17213886 | EGR2 | 1959 | 4.061E-6 | trans | ||
| rs7200513 | chr16 | 17219474 | EGR2 | 1959 | 0.01 | trans | ||
| rs3829495 | chr16 | 17221986 | EGR2 | 1959 | 5.763E-4 | trans | ||
| rs7193763 | chr16 | 17225534 | EGR2 | 1959 | 5.351E-5 | trans | ||
| rs11861635 | chr16 | 82543377 | EGR2 | 1959 | 0.03 | trans | ||
| rs276966 | chr16 | 86233753 | EGR2 | 1959 | 0.03 | trans | ||
| rs643099 | chr17 | 6568815 | EGR2 | 1959 | 0.19 | trans | ||
| rs1867231 | chr17 | 13678834 | EGR2 | 1959 | 0.06 | trans | ||
| rs9903086 | chr17 | 39949515 | EGR2 | 1959 | 0.16 | trans | ||
| rs7219986 | chr17 | 53263206 | EGR2 | 1959 | 0 | trans | ||
| rs17817797 | chr17 | 53332496 | EGR2 | 1959 | 9.358E-4 | trans | ||
| rs16951770 | chr18 | 7444826 | EGR2 | 1959 | 0.01 | trans | ||
| rs2851752 | chr18 | 25350684 | EGR2 | 1959 | 0.11 | trans | ||
| rs7244394 | chr18 | 32223105 | EGR2 | 1959 | 0.02 | trans | ||
| snp_a-4248697 | 0 | EGR2 | 1959 | 0.02 | trans | |||
| rs16951384 | chr18 | 47527116 | EGR2 | 1959 | 0.08 | trans | ||
| rs10502924 | chr18 | 49172178 | EGR2 | 1959 | 0.12 | trans | ||
| rs17064695 | chr18 | 55956427 | EGR2 | 1959 | 0.05 | trans | ||
| rs1469489 | chr18 | 57228488 | EGR2 | 1959 | 0.15 | trans | ||
| rs1469490 | chr18 | 57229886 | EGR2 | 1959 | 0.11 | trans | ||
| rs17074993 | chr18 | 63260128 | EGR2 | 1959 | 0.01 | trans | ||
| snp_a-1821751 | 0 | EGR2 | 1959 | 0 | trans | |||
| rs1942461 | chr18 | 70509159 | EGR2 | 1959 | 0.01 | trans | ||
| rs1942462 | chr18 | 70509823 | EGR2 | 1959 | 0.01 | trans | ||
| rs1942463 | chr18 | 70509841 | EGR2 | 1959 | 0.01 | trans | ||
| rs1942464 | chr18 | 70509877 | EGR2 | 1959 | 2.489E-4 | trans | ||
| rs1942466 | chr18 | 70517154 | EGR2 | 1959 | 0.01 | trans | ||
| rs2156494 | chr18 | 70518084 | EGR2 | 1959 | 0.01 | trans | ||
| rs17056922 | chr18 | 73086804 | EGR2 | 1959 | 0.01 | trans | ||
| rs1540047 | chr18 | 75047028 | EGR2 | 1959 | 0.07 | trans | ||
| rs2850843 | chr18 | 75048905 | EGR2 | 1959 | 0.07 | trans | ||
| rs1540051 | chr18 | 75049934 | EGR2 | 1959 | 0.02 | trans | ||
| rs16980096 | chr19 | 15000921 | EGR2 | 1959 | 0.07 | trans | ||
| snp_a-2138575 | 0 | EGR2 | 1959 | 0 | trans | |||
| rs8103530 | chr19 | 39172219 | EGR2 | 1959 | 0.13 | trans | ||
| rs234337 | chr19 | 40458724 | EGR2 | 1959 | 0 | trans | ||
| rs234363 | chr19 | 40474607 | EGR2 | 1959 | 5.133E-4 | trans | ||
| rs634346 | chr20 | 3587203 | EGR2 | 1959 | 0.19 | trans | ||
| rs6417634 | chr20 | 6408617 | EGR2 | 1959 | 0.15 | trans | ||
| rs6085552 | chr20 | 6409004 | EGR2 | 1959 | 0.15 | trans | ||
| rs16997215 | chr20 | 16101890 | EGR2 | 1959 | 0.02 | trans | ||
| snp_a-4254460 | 0 | EGR2 | 1959 | 0.02 | trans | |||
| rs8113962 | chr20 | 18995999 | EGR2 | 1959 | 0.13 | trans | ||
| rs1797040 | chr20 | 23907743 | EGR2 | 1959 | 0.01 | trans | ||
| rs2424603 | 0 | EGR2 | 1959 | 0.01 | trans | |||
| rs6102879 | chr20 | 41138766 | EGR2 | 1959 | 0.02 | trans | ||
| rs6026122 | chr20 | 56769083 | EGR2 | 1959 | 7.113E-4 | trans | ||
| rs259991 | chr20 | 57728347 | EGR2 | 1959 | 0.06 | trans | ||
| rs6015452 | chr20 | 57782312 | EGR2 | 1959 | 0.04 | trans | ||
| rs6124034 | chr20 | 59517206 | EGR2 | 1959 | 0.04 | trans | ||
| rs6124036 | chr20 | 59517478 | EGR2 | 1959 | 0.04 | trans | ||
| rs4812237 | chr20 | 59518040 | EGR2 | 1959 | 0.01 | trans | ||
| rs2847443 | chr21 | 14595263 | EGR2 | 1959 | 0.05 | trans | ||
| rs2226440 | chr21 | 18066174 | EGR2 | 1959 | 0.1 | trans | ||
| rs16992859 | chr21 | 36507007 | EGR2 | 1959 | 0.02 | trans | ||
| rs2834872 | chr21 | 36634845 | EGR2 | 1959 | 0.01 | trans | ||
| rs2834874 | chr21 | 36641270 | EGR2 | 1959 | 0.04 | trans | ||
| rs2834875 | chr21 | 36656119 | EGR2 | 1959 | 0 | trans | ||
| rs877910 | chr21 | 36658376 | EGR2 | 1959 | 0.01 | trans | ||
| rs7284012 | chr21 | 36664376 | EGR2 | 1959 | 0.07 | trans | ||
| rs6586271 | chr21 | 44362641 | EGR2 | 1959 | 0.03 | trans | ||
| rs369613 | chr22 | 22380717 | EGR2 | 1959 | 0.06 | trans | ||
| rs410285 | chr22 | 22381767 | EGR2 | 1959 | 0.06 | trans | ||
| rs134028 | chr22 | 28048448 | EGR2 | 1959 | 1.896E-5 | trans | ||
| rs5762317 | chr22 | 28094988 | EGR2 | 1959 | 0 | trans | ||
| rs9968025 | chr22 | 32321588 | EGR2 | 1959 | 0.04 | trans | ||
| rs5768165 | chr22 | 48318197 | EGR2 | 1959 | 0.03 | trans | ||
| rs5768168 | chr22 | 48319334 | EGR2 | 1959 | 0.03 | trans | ||
| rs5768173 | chr22 | 48321475 | EGR2 | 1959 | 0.03 | trans | ||
| rs16978981 | chrX | 13538144 | EGR2 | 1959 | 0.19 | trans | ||
| rs7879159 | chrX | 19481591 | EGR2 | 1959 | 0.1 | trans | ||
| rs12006821 | chrX | 50424687 | EGR2 | 1959 | 0.02 | trans | ||
| rs5977652 | chrX | 131315199 | EGR2 | 1959 | 0.15 | trans | ||
| rs16992781 | chrX | 146460376 | EGR2 | 1959 | 0.02 | trans | ||
| rs4824295 | chrX | 146496881 | EGR2 | 1959 | 0.01 | trans |
Section II. Transcriptome annotation
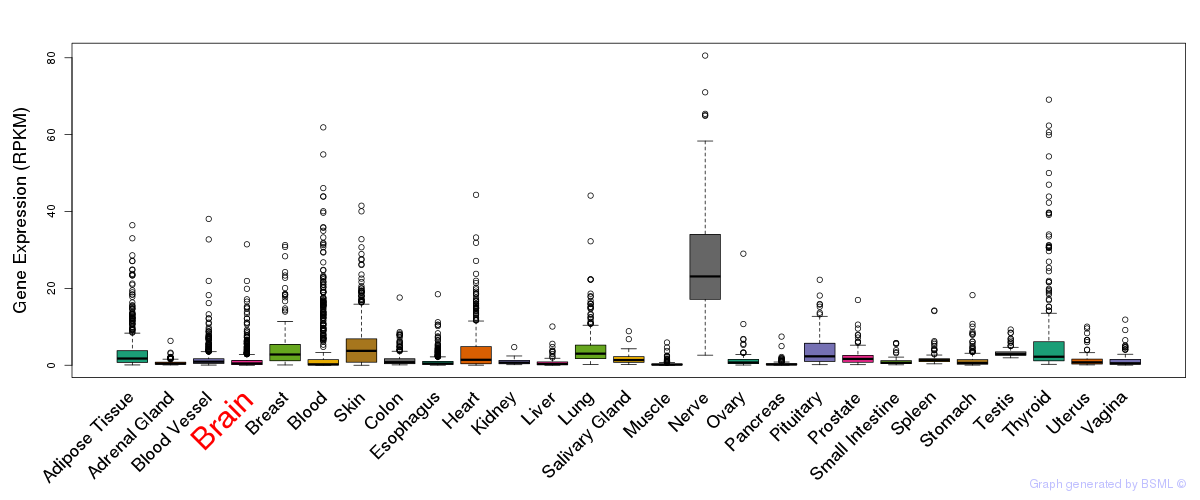
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PMPCB | 0.89 | 0.85 |
| UQCRC2 | 0.88 | 0.85 |
| DNAJA2 | 0.88 | 0.84 |
| YPEL5 | 0.88 | 0.87 |
| POLR3F | 0.87 | 0.85 |
| BTBD10 | 0.87 | 0.87 |
| SARS | 0.87 | 0.86 |
| TMEM49 | 0.87 | 0.84 |
| COPS3 | 0.86 | 0.83 |
| RHOT1 | 0.86 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.67 | -0.59 |
| AF347015.26 | -0.67 | -0.59 |
| MT-CO2 | -0.66 | -0.57 |
| AF347015.8 | -0.65 | -0.57 |
| MT-CYB | -0.65 | -0.57 |
| AF347015.15 | -0.64 | -0.57 |
| AF347015.33 | -0.63 | -0.57 |
| AF347015.21 | -0.63 | -0.55 |
| AF347015.18 | -0.63 | -0.63 |
| AF347015.31 | -0.62 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 14532282 | |
| GO:0005515 | protein binding | IPI | 14532282 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | TAS | Brain (GO term level: 7) | 9537424 |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007422 | peripheral nervous system development | TAS | 9537424 | |
| GO:0007638 | mechanosensory behavior | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GESERICK TERT TARGETS DN | 21 | 16 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| MCBRYAN TERMINAL END BUD UP | 12 | 12 | All SZGR 2.0 genes in this pathway |
| CHASSOT SKIN WOUND | 10 | 9 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| STAEGE EWING FAMILY TUMOR | 33 | 22 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE UP | 34 | 27 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 30MIN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES | 31 | 26 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN DEDIFFERENTIATED STATE DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| UZONYI RESPONSE TO LEUKOTRIENE AND THROMBIN | 37 | 26 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 1HR UP | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| PHONG TNF TARGETS UP | 63 | 43 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
| HECKER IFNB1 TARGETS | 95 | 54 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 907 | 913 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 858 | 864 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-137 | 579 | 586 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| miR-141/200a | 1029 | 1035 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-3p | 73 | 79 | m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-150 | 401 | 408 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-17-5p/20/93.mr/106/519.d | 458 | 465 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-224 | 1019 | 1026 | 1A,m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-25/32/92/363/367 | 612 | 618 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-324-5p | 826 | 832 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-34b | 962 | 968 | 1A | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-377 | 864 | 870 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.