Gene Page: ELK1
Summary ?
| GeneID | 2002 |
| Symbol | ELK1 |
| Synonyms | - |
| Description | ELK1, ETS transcription factor |
| Reference | MIM:311040|HGNC:HGNC:3321|Ensembl:ENSG00000126767|HPRD:02407|Vega:OTTHUMG00000021452 |
| Gene type | protein-coding |
| Map location | Xp11.2 |
| Sherlock p-value | 0.234 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.162:DS1_beta=0.019600:DS2_p=3.20e-01:DS2_beta=0.050:DS2_FDR=5.73e-01 |
| Fetal beta | -0.472 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.879 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
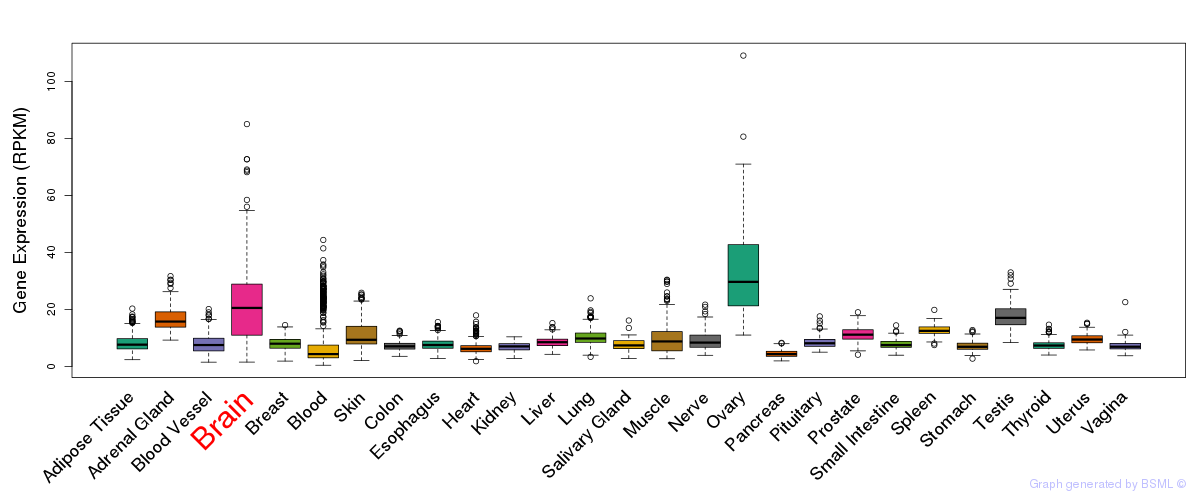
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMG5 | 0.87 | 0.88 |
| FLII | 0.85 | 0.86 |
| GRIPAP1 | 0.85 | 0.85 |
| RECQL5 | 0.84 | 0.85 |
| XPC | 0.84 | 0.85 |
| FAM160B2 | 0.84 | 0.86 |
| MYBBP1A | 0.84 | 0.85 |
| FKBP15 | 0.84 | 0.84 |
| DDX27 | 0.83 | 0.85 |
| POLG | 0.83 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.70 | -0.68 |
| AF347015.31 | -0.65 | -0.60 |
| C1orf54 | -0.64 | -0.62 |
| CLEC2B | -0.61 | -0.62 |
| MT-CO2 | -0.60 | -0.56 |
| GNG11 | -0.59 | -0.59 |
| AF347015.27 | -0.58 | -0.56 |
| IFI27 | -0.58 | -0.53 |
| VAMP5 | -0.58 | -0.57 |
| HIGD1B | -0.58 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 14970218 | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 16291755 |16533805 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 14970218 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IC | 14970218 | |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 11313879 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD | 1547942 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western Phenotypic Enhancement Reconstituted Complex | BioGRID | 12514134 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Reconstituted Complex | BioGRID | 9010223 |
| FLI1 | EWSR2 | SIC-1 | Friend leukemia virus integration 1 | Reconstituted Complex | BioGRID | 9010223 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 7540136 |
| FOSL1 | FRA | FRA1 | fra-1 | FOS-like antigen 1 | Elk1 interacts with the region of the FRA-1 promoter containing the SRE and ATF sites. | BIND | 15806162 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 8798570 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 8798570 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Mapk1 (p42Mapk) phosphorylates Elk1 at Ser383. | BIND | 15782123 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD | 8208531 |8586671 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Biochemical Activity Reconstituted Complex | BioGRID | 8586671 |12594221 |
| MAPK11 | P38B | P38BETA2 | PRKM11 | SAPK2 | SAPK2B | p38-2 | p38Beta | mitogen-activated protein kinase 11 | p38-2 interacts with and phosphorylates Elk1. | BIND | 9235954 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | Elk-1 is phosphorylated by an unspecified isoform of p38 MAPK. This interaction was modeled on a demonstrated interaction between human Elk-1 and p38 MAPK from an unspecified species. | BIND | 9130707 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD | 8622669 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | An unspecified isoform of p38 interacts with and phosphorylates Elk-1. | BIND | 9155018 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Elk-1 is phosphorylated by ERK-1. This interaction was modeled on a demonstrated interaction between human Elk-1 and ERK-1 from an unspecified species. | BIND | 9130707 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 8208531 |12473660 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Mapk3 (p44Mapk) phosphorylates Elk1 at Ser383. | BIND | 15782123 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | ERK1 interacts with and phosphorylates Elk1. This interaction was modelled on a demonstrated interaction between rat ERK1 and human Elk1. | BIND | 9235954 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Elk-1 is phosphorylated by ERK-1. This interaction was modeled on a demonstrated interaction between human Elk-1 and ERK-1 from an unspecified species. | BIND | 9020136 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | ERK1 interacts with and phosphorylates Elk-1. | BIND | 9155018 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Erk1 phosphorylates Elk-1. This interaction was modeled on a demonstrated interaction between Erk1 and Elk-1 from an unspecified species. | BIND | 10431817 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | An unspecified isoform of JNK1 interacts with and phosphorylates Elk1. | BIND | 9235954 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Biochemical Activity | BioGRID | 8586671 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Elk-1 is phosphorylated by JNK-1. This interaction was modeled on a demonstrated interaction between human Elk-1 and JNK-1 from an unspecified species. | BIND | 9020136 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | An unspecified isoform of JNK2 interacts with and phosphorylates Elk-1. | BIND | 9155018 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | ELK1 interacts with PIAS2a (PIASX-alpha). | BIND | 15920481 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 11279280 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD,BioGRID | 9010223 |
| STK16 | FLJ39635 | KRCT | MPSK | PKL12 | TSF1 | serine/threonine kinase 16 | - | HPRD,BioGRID | 10947953 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGF PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPO PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1 PATHWAY | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL6 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INSULIN PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NGF PATHWAY | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PDGF PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| SA TRKA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK1 AND MAPK2 TARGETS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK8 AND MAPK9 TARGETS | 8 | 7 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK11 TARGETS | 5 | 5 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK14 TARGETS | 10 | 10 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C4 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER DN | 33 | 22 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES | 31 | 26 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM HIGH RISK UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 678 | 684 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-136 | 729 | 735 | m8 | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-150 | 541 | 547 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG | ||||
| miR-185 | 49 | 56 | 1A,m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-219 | 365 | 371 | m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-326 | 1251 | 1258 | 1A,m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-382 | 191 | 197 | m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-539 | 405 | 412 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.