Gene Page: FLCN
Summary ?
| GeneID | 201163 |
| Symbol | FLCN |
| Synonyms | BHD|FLCL |
| Description | folliculin |
| Reference | MIM:607273|HGNC:HGNC:27310|Ensembl:ENSG00000154803|HPRD:06278|Vega:OTTHUMG00000059275 |
| Gene type | protein-coding |
| Map location | 17p11.2 |
| Pascal p-value | 0.162 |
| Sherlock p-value | 0.844 |
| DEG p-value | DEG:Zhao_2015:p=5.05e-05:q=0.0489 |
| Fetal beta | -0.546 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8065836 | chr17 | 16837364 | FLCN | 201163 | 0.15 | cis | ||
| rs3803761 | chr17 | 17116411 | FLCN | 201163 | 0.15 | cis | ||
| rs7010362 | chr8 | 23218100 | FLCN | 201163 | 0.17 | trans | ||
| rs2701130 | chr12 | 52429439 | FLCN | 201163 | 0.03 | trans | ||
| rs3899870 | chr13 | 97205717 | FLCN | 201163 | 0.01 | trans | ||
| rs9908205 | 17 | 17105615 | FLCN | ENSG00000154803.8 | 1.916E-6 | 0 | 26428 | gtex_brain_putamen_basal |
| rs9908150 | 17 | 17114061 | FLCN | ENSG00000154803.8 | 2.774E-7 | 0 | 17982 | gtex_brain_putamen_basal |
| rs4399589 | 17 | 17114586 | FLCN | ENSG00000154803.8 | 2.834E-6 | 0 | 17457 | gtex_brain_putamen_basal |
| rs7218795 | 17 | 17115566 | FLCN | ENSG00000154803.8 | 2.834E-6 | 0 | 16477 | gtex_brain_putamen_basal |
| rs3803761 | 17 | 17116412 | FLCN | ENSG00000154803.8 | 1.909E-6 | 0 | 15631 | gtex_brain_putamen_basal |
| rs7208065 | 17 | 17120204 | FLCN | ENSG00000154803.8 | 1.115E-8 | 0 | 11839 | gtex_brain_putamen_basal |
| rs4985705 | 17 | 17120668 | FLCN | ENSG00000154803.8 | 4.575E-9 | 0 | 11375 | gtex_brain_putamen_basal |
| rs4985752 | 17 | 17120812 | FLCN | ENSG00000154803.8 | 1.17E-7 | 0 | 11231 | gtex_brain_putamen_basal |
| rs8080386 | 17 | 17121624 | FLCN | ENSG00000154803.8 | 7.52E-8 | 0 | 10419 | gtex_brain_putamen_basal |
| rs1736221 | 17 | 17125595 | FLCN | ENSG00000154803.8 | 7.52E-8 | 0 | 6448 | gtex_brain_putamen_basal |
| rs1736220 | 17 | 17126792 | FLCN | ENSG00000154803.8 | 7.52E-8 | 0 | 5251 | gtex_brain_putamen_basal |
| rs1708618 | 17 | 17131869 | FLCN | ENSG00000154803.8 | 7.532E-10 | 0 | 174 | gtex_brain_putamen_basal |
| rs1736215 | 17 | 17132202 | FLCN | ENSG00000154803.8 | 2.368E-9 | 0 | -159 | gtex_brain_putamen_basal |
| rs1736214 | 17 | 17132333 | FLCN | ENSG00000154803.8 | 3.22E-9 | 0 | -290 | gtex_brain_putamen_basal |
| rs66460366 | 17 | 17132818 | FLCN | ENSG00000154803.8 | 7.079E-8 | 0 | -775 | gtex_brain_putamen_basal |
| rs1613416 | 17 | 17133121 | FLCN | ENSG00000154803.8 | 3.551E-9 | 0 | -1078 | gtex_brain_putamen_basal |
| rs1736213 | 17 | 17134528 | FLCN | ENSG00000154803.8 | 9.897E-9 | 0 | -2485 | gtex_brain_putamen_basal |
| rs10459910 | 17 | 17136891 | FLCN | ENSG00000154803.8 | 1.523E-7 | 0 | -4848 | gtex_brain_putamen_basal |
| rs1708629 | 17 | 17140297 | FLCN | ENSG00000154803.8 | 9.197E-8 | 0 | -8254 | gtex_brain_putamen_basal |
| rs1736207 | 17 | 17144854 | FLCN | ENSG00000154803.8 | 3.528E-7 | 0 | -12811 | gtex_brain_putamen_basal |
| rs1708626 | 17 | 17144908 | FLCN | ENSG00000154803.8 | 3.69E-7 | 0 | -12865 | gtex_brain_putamen_basal |
| rs1736206 | 17 | 17144978 | FLCN | ENSG00000154803.8 | 3.69E-7 | 0 | -12935 | gtex_brain_putamen_basal |
| rs1624963 | 17 | 17146368 | FLCN | ENSG00000154803.8 | 1.641E-8 | 0 | -14325 | gtex_brain_putamen_basal |
| rs1708627 | 17 | 17146959 | FLCN | ENSG00000154803.8 | 3.395E-6 | 0 | -14916 | gtex_brain_putamen_basal |
| rs1736203 | 17 | 17151664 | FLCN | ENSG00000154803.8 | 2.792E-9 | 0 | -19621 | gtex_brain_putamen_basal |
| rs1736202 | 17 | 17151938 | FLCN | ENSG00000154803.8 | 1.436E-9 | 0 | -19895 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
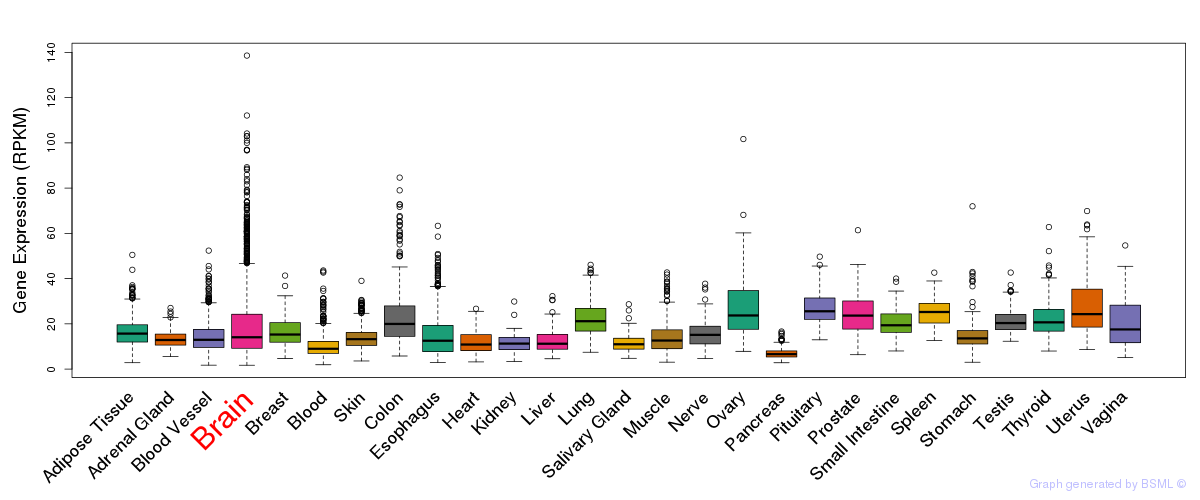
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |