Gene Page: ENO2
Summary ?
| GeneID | 2026 |
| Symbol | ENO2 |
| Synonyms | HEL-S-279|NSE |
| Description | enolase 2 (gamma, neuronal) |
| Reference | MIM:131360|HGNC:HGNC:3353|Ensembl:ENSG00000111674|HPRD:00573|Vega:OTTHUMG00000168967 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.466 |
| Sherlock p-value | 0.302 |
| Fetal beta | -1.446 |
| Support | CELL METABOLISM G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
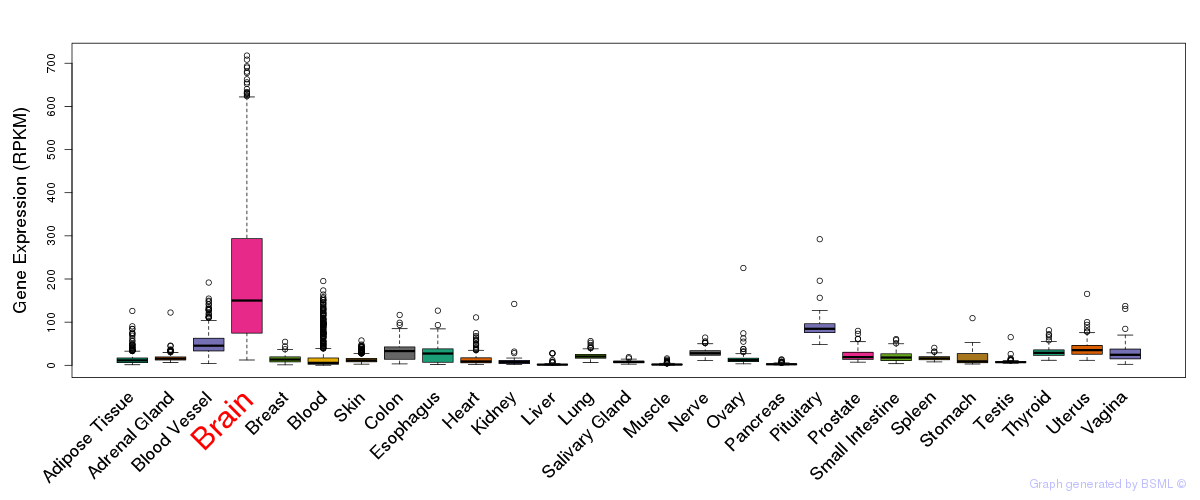
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FOXP2 | 0.88 | 0.42 |
| LHX9 | 0.87 | 0.19 |
| FIGN | 0.87 | 0.34 |
| TRPM8 | 0.85 | 0.31 |
| TCF7L2 | 0.85 | 0.22 |
| ANO2 | 0.84 | 0.26 |
| GBX2 | 0.84 | 0.25 |
| FAM83G | 0.84 | 0.31 |
| TRH | 0.83 | 0.19 |
| MAP4K1 | 0.81 | 0.23 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTDC1 | -0.20 | 0.09 |
| JAM2 | -0.20 | 0.02 |
| RGS20 | -0.20 | -0.28 |
| PIK3IP1 | -0.20 | 0.04 |
| COX4I2 | -0.20 | -0.32 |
| AIF1L | -0.20 | -0.23 |
| CKB | -0.19 | 0.02 |
| PPP2R5A | -0.19 | -0.02 |
| SLC26A4 | -0.19 | 0.02 |
| ARSF | -0.19 | -0.19 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004634 | phosphopyruvate hydratase activity | TAS | 2045099 | |
| GO:0016829 | lyase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006096 | glycolysis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000015 | phosphopyruvate hydratase complex | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANXA11 | ANX11 | CAP50 | annexin A11 | Two-hybrid | BioGRID | 16169070 |
| C16orf45 | FLJ32618 | chromosome 16 open reading frame 45 | Two-hybrid | BioGRID | 16169070 |
| C6orf47 | D6S53E | G4 | NG34 | chromosome 6 open reading frame 47 | Two-hybrid | BioGRID | 16169070 |
| ENSA | MGC4319 | MGC78563 | MGC8394 | endosulfine alpha | Two-hybrid | BioGRID | 16169070 |
| HABP4 | IHABP4 | Ki-1/57 | MGC825 | SERBP1L | hyaluronan binding protein 4 | Two-hybrid | BioGRID | 16169070 |
| HK1 | HK1-ta | HK1-tb | HK1-tc | HKI | HXK1 | hexokinase 1 | Two-hybrid | BioGRID | 16169070 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | Two-hybrid | BioGRID | 16169070 |
| IL17B | IL-17B | IL-20 | MGC138900 | MGC138901 | ZCYTO7 | interleukin 17B | Two-hybrid | BioGRID | 16169070 |
| MAP4 | DKFZp779A1753 | MGC8617 | microtubule-associated protein 4 | Two-hybrid | BioGRID | 16169070 |
| NAT9 | DKFZp564C103 | EBSP | N-acetyltransferase 9 (GCN5-related, putative) | Two-hybrid | BioGRID | 16169070 |
| NDUFS7 | CI-20KD | FLJ45860 | FLJ46880 | MGC120002 | MY017 | PSST | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | Two-hybrid | BioGRID | 16169070 |
| SIGIRR | MGC110992 | TIR8 | single immunoglobulin and toll-interleukin 1 receptor (TIR) domain | Two-hybrid | BioGRID | 16169070 |
| SNUPN | KPNBL | RNUT1 | Snurportin1 | snurportin 1 | Two-hybrid | BioGRID | 16169070 |
| ST3GAL2 | Gal-NAc6S | SIAT4B | ST3GALII | ST3GalA.2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | Two-hybrid | BioGRID | 16169070 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | Two-hybrid | BioGRID | 16169070 |
| UBE2C | UBCH10 | dJ447F3.2 | ubiquitin-conjugating enzyme E2C | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCONEOGENESIS | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONAL CARCINOMA UP | 41 | 27 | All SZGR 2.0 genes in this pathway |
| KORKOLA SEMINOMA UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| TOMIDA METASTASIS DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| MAINA VHL TARGETS DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| WOTTON RUNX TARGETS DN | 29 | 18 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA MAST CELL | 46 | 34 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 DN | 52 | 35 | All SZGR 2.0 genes in this pathway |
| KIM HYPOXIA | 25 | 21 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA UP | 28 | 23 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 UP | 87 | 50 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOLYSIS | 21 | 13 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| WACKER HYPOXIA TARGETS OF VHL | 13 | 11 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |