Gene Page: C2orf69
Summary ?
| GeneID | 205327 |
| Symbol | C2orf69 |
| Synonyms | - |
| Description | chromosome 2 open reading frame 69 |
| Reference | HGNC:HGNC:26799|Ensembl:ENSG00000178074|HPRD:08797|Vega:OTTHUMG00000154480 |
| Gene type | protein-coding |
| Map location | 2q33.1 |
| Pascal p-value | 3.334E-12 |
| Fetal beta | -0.03 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18202627 | 2 | 200776533 | C2orf69 | 3.2E-8 | -0.009 | 9.65E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs823123 | chr1 | 205725345 | C2orf69 | 205327 | 0.13 | trans | ||
| rs823066 | chr1 | 205771172 | C2orf69 | 205327 | 0 | trans | ||
| rs11120206 | chr1 | 206610593 | C2orf69 | 205327 | 0.16 | trans | ||
| rs2716734 | chr2 | 39947720 | C2orf69 | 205327 | 0.01 | trans | ||
| rs2716736 | chr2 | 39947946 | C2orf69 | 205327 | 0.01 | trans | ||
| rs6770074 | chr3 | 5496504 | C2orf69 | 205327 | 0.11 | trans | ||
| rs9882137 | chr3 | 29494146 | C2orf69 | 205327 | 0.01 | trans | ||
| rs9882501 | chr3 | 29494429 | C2orf69 | 205327 | 0.1 | trans | ||
| rs467856 | chr5 | 3128168 | C2orf69 | 205327 | 0.13 | trans | ||
| snp_a-2169623 | 0 | C2orf69 | 205327 | 4.897E-5 | trans | |||
| rs13175086 | chr5 | 174691428 | C2orf69 | 205327 | 0.01 | trans | ||
| rs16894557 | chr6 | 28999825 | C2orf69 | 205327 | 0.05 | trans | ||
| rs2016128 | chr6 | 42442743 | C2orf69 | 205327 | 0.04 | trans | ||
| rs4947631 | chr7 | 50603378 | C2orf69 | 205327 | 0.02 | trans | ||
| rs17139868 | chr7 | 117079228 | C2orf69 | 205327 | 0.09 | trans | ||
| rs2195588 | chr8 | 40356699 | C2orf69 | 205327 | 0.18 | trans | ||
| rs10124277 | chr9 | 8499294 | C2orf69 | 205327 | 0.08 | trans | ||
| rs12352894 | chr9 | 38197383 | C2orf69 | 205327 | 0.18 | trans | ||
| rs6559219 | 0 | C2orf69 | 205327 | 0.18 | trans | |||
| rs12255258 | chr10 | 106306927 | C2orf69 | 205327 | 0.01 | trans | ||
| rs998923 | chr13 | 35507810 | C2orf69 | 205327 | 0.04 | trans | ||
| rs4943304 | chr13 | 36058065 | C2orf69 | 205327 | 0.19 | trans | ||
| rs1503026 | chr18 | 70842605 | C2orf69 | 205327 | 0.11 | trans | ||
| rs12326929 | chr18 | 70864592 | C2orf69 | 205327 | 0.1 | trans | ||
| rs12327665 | chr19 | 39622639 | C2orf69 | 205327 | 0.1 | trans | ||
| rs6040607 | chr20 | 11371092 | C2orf69 | 205327 | 0.04 | trans | ||
| rs225434 | chr21 | 43724740 | C2orf69 | 205327 | 0.1 | trans |
Section II. Transcriptome annotation
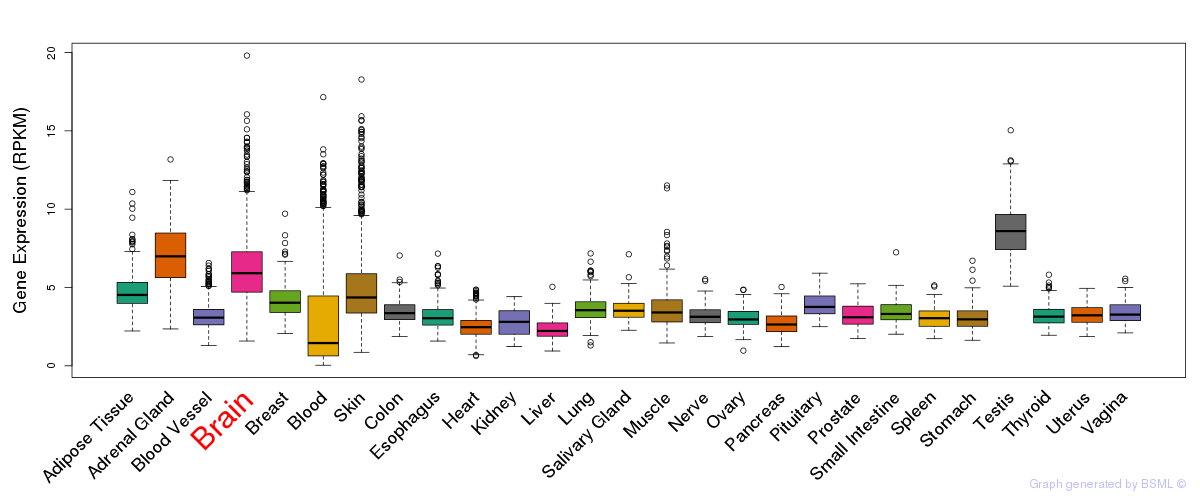
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |