Gene Page: EYA1
Summary ?
| GeneID | 2138 |
| Symbol | EYA1 |
| Synonyms | BOP|BOR|BOS1|OFC1 |
| Description | EYA transcriptional coactivator and phosphatase 1 |
| Reference | MIM:601653|HGNC:HGNC:3519|Ensembl:ENSG00000104313|HPRD:03388|Vega:OTTHUMG00000149894 |
| Gene type | protein-coding |
| Map location | 8q13.3 |
| Pascal p-value | 0.53 |
| Fetal beta | 0.387 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2890901 | chr9 | 10112765 | EYA1 | 2138 | 0.17 | trans | ||
| rs10958892 | chr9 | 10125117 | EYA1 | 2138 | 0.01 | trans | ||
| rs10958896 | chr9 | 10126894 | EYA1 | 2138 | 0.01 | trans | ||
| rs10958899 | chr9 | 10128700 | EYA1 | 2138 | 0.01 | trans | ||
| rs10958907 | chr9 | 10131580 | EYA1 | 2138 | 0 | trans | ||
| rs1322159 | chr9 | 10132695 | EYA1 | 2138 | 0.12 | trans | ||
| rs10958910 | chr9 | 10134759 | EYA1 | 2138 | 0.03 | trans |
Section II. Transcriptome annotation
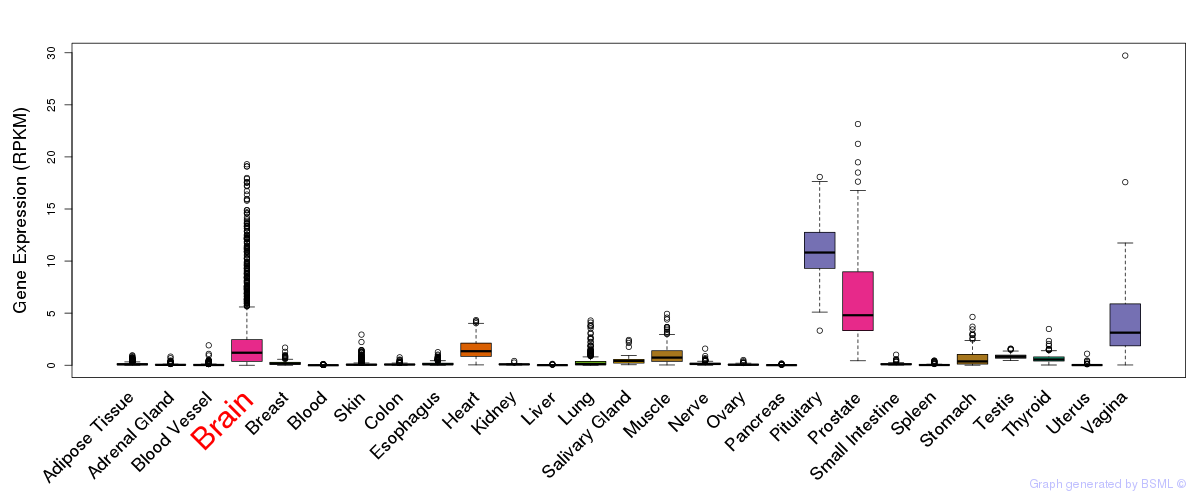
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRLR | 0.97 | 0.15 |
| CP | 0.97 | 0.62 |
| KCNJ13 | 0.96 | 0.28 |
| CLIC6 | 0.95 | 0.47 |
| SLC5A5 | 0.94 | 0.38 |
| SLC13A4 | 0.91 | 0.30 |
| ABCA4 | 0.90 | 0.40 |
| FOLR1 | 0.88 | 0.33 |
| RBM47 | 0.87 | 0.16 |
| TRPV4 | 0.87 | 0.22 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.12 | -0.54 |
| OLFM2 | -0.12 | -0.43 |
| PLEKHO1 | -0.12 | -0.52 |
| ALKBH2 | -0.12 | -0.54 |
| ASGR1 | -0.12 | -0.41 |
| STMN1 | -0.12 | -0.45 |
| AC009133.1 | -0.11 | -0.46 |
| RFC2 | -0.11 | -0.45 |
| TUBB2B | -0.11 | -0.53 |
| MPND | -0.11 | -0.35 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004725 | protein tyrosine phosphatase activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045664 | regulation of neuron differentiation | IEA | neuron, neurogenesis (GO term level: 9) | - |
| GO:0001657 | ureteric bud development | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 9020840 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007605 | sensory perception of sound | TAS | 9020840 | |
| GO:0007389 | pattern specification process | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0042472 | inner ear morphogenesis | IEA | - | |
| GO:0045165 | cell fate commitment | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR | 27 | 12 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1810 | 1816 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| hsa-miR-101 | UACAGUACUGUGAUAACUGAAG | ||||
| hsa-miR-101 | UACAGUACUGUGAUAACUGAAG | ||||
| miR-124.1 | 849 | 855 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 848 | 855 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 200 | 206 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-144 | 1810 | 1816 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-15/16/195/424/497 | 1646 | 1653 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-219 | 705 | 712 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-23 | 326 | 332 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-27 | 282 | 288 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC | ||||
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-323 | 326 | 332 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-369-3p | 1081 | 1088 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1082 | 1088 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-493-5p | 691 | 697 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-503 | 1647 | 1653 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-543 | 229 | 235 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.