Gene Page: PUF60
Summary ?
| GeneID | 22827 |
| Symbol | PUF60 |
| Synonyms | FIR|RoBPI|SIAHBP1|VRJS |
| Description | poly(U) binding splicing factor 60KDa |
| Reference | MIM:604819|HGNC:HGNC:17042|Ensembl:ENSG00000179950|HPRD:18051|Vega:OTTHUMG00000165155 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 7.597E-5 |
| Sherlock p-value | 0.208 |
| TADA p-value | 0.012 |
| Fetal beta | 0.584 |
| DMG | 1 (# studies) |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PUF60 | chr8 | 144898899 | C | T | NM_001136033 NM_001271096 NM_001271097 NM_001271098 NM_001271099 NM_001271100 NM_014281 NM_078480 | p.448G>R p.473G>R p.445G>R p.490G>R p.462G>R p.431G>R p.474G>R p.491G>R | missense missense missense missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18431951 | 8 | 144906507 | PUF60 | 1.39E-4 | 0.008 | 0.136 | DMG:Montano_2016 |
Section II. Transcriptome annotation
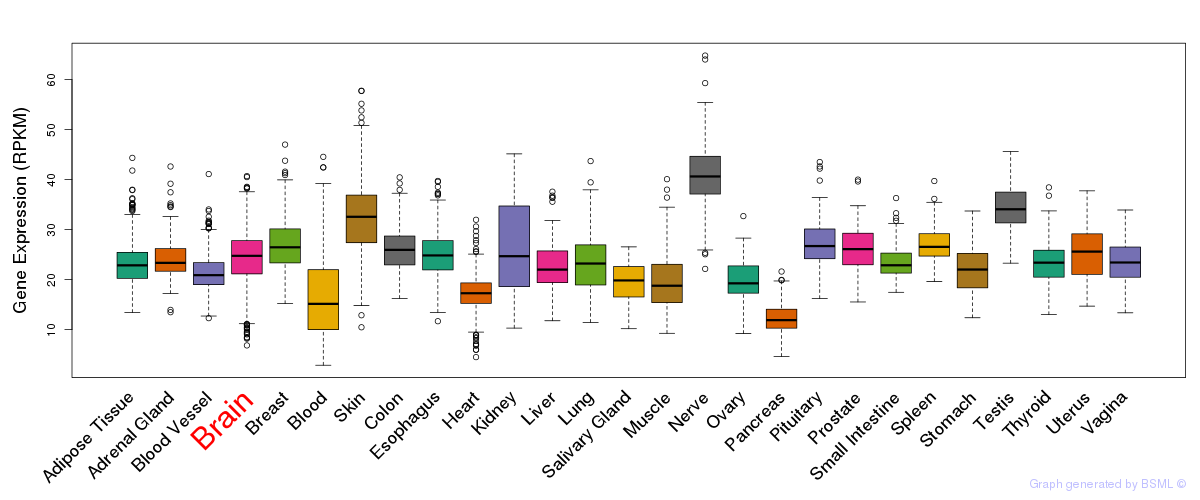
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NMNAT2 | 0.91 | 0.93 |
| CLCN4 | 0.91 | 0.91 |
| FOXO3 | 0.90 | 0.92 |
| HERC1 | 0.89 | 0.92 |
| ATCAY | 0.89 | 0.92 |
| PRDM2 | 0.89 | 0.92 |
| GSK3B | 0.88 | 0.93 |
| SMURF1 | 0.88 | 0.91 |
| RNF111 | 0.88 | 0.91 |
| UBE2O | 0.88 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.71 | -0.78 |
| SERPINB6 | -0.69 | -0.73 |
| AF347015.31 | -0.69 | -0.76 |
| S100A16 | -0.69 | -0.75 |
| HIGD1B | -0.69 | -0.78 |
| MT-CO2 | -0.68 | -0.77 |
| TLCD1 | -0.68 | -0.75 |
| AC021016.1 | -0.67 | -0.74 |
| COPZ2 | -0.66 | -0.71 |
| AF347015.33 | -0.66 | -0.72 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD2BP2 | FWP010 | LIN1 | Snu40 | U5-52K | CD2 (cytoplasmic tail) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| DDX3X | DBX | DDX14 | DDX3 | HLP2 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | Affinity Capture-MS | BioGRID | 17353931 |
| ELAC2 | ELC2 | FLJ10530 | FLJ36693 | FLJ42848 | HPC2 | elaC homolog 2 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | - | HPRD | 10882074 |
| G3BP1 | G3BP | HDH-VIII | MGC111040 | GTPase activating protein (SH3 domain) binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPA3 | 2610510D13Rik | D10S102 | FBRNP | HNRPA3 | MGC138232 | MGC142030 | heterogeneous nuclear ribonucleoprotein A3 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPL | FLJ35509 | HNRPL | P/OKcl.14 | hnRNP-L | heterogeneous nuclear ribonucleoprotein L | Affinity Capture-MS | BioGRID | 17353931 |
| IGF2BP1 | CRD-BP | CRDBP | IMP-1 | IMP1 | VICKZ1 | ZBP1 | insulin-like growth factor 2 mRNA binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| IGF2BP2 | IMP-2 | IMP2 | VICKZ2 | p62 | insulin-like growth factor 2 mRNA binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| IGF2BP3 | DKFZp686F1078 | IMP-3 | IMP3 | KOC1 | VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 | Affinity Capture-MS | BioGRID | 17353931 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Affinity Capture-MS | BioGRID | 17353931 |
| KHSRP | FBP2 | FUBP2 | KSRP | MGC99676 | KH-type splicing regulatory protein | Affinity Capture-MS | BioGRID | 17353931 |
| LOH12CR1 | LOH1CR12 | loss of heterozygosity, 12, chromosomal region 1 | Two-hybrid | BioGRID | 16189514 |
| MOV10 | DKFZp667O1423 | FLJ32791 | KIAA1631 | fSAP113 | gb110 | Mov10, Moloney leukemia virus 10, homolog (mouse) | Affinity Capture-MS | BioGRID | 17353931 |
| OLA1 | DKFZp313H1942 | GTPBP9 | PTD004 | Obg-like ATPase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| P4HB | DSI | ERBA2L | GIT | PDI | PDIA1 | PHDB | PO4DB | PO4HB | PROHB | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| PABPC1 | PAB1 | PABP | PABP1 | PABPC2 | PABPL1 | poly(A) binding protein, cytoplasmic 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PABPC4 | APP-1 | APP1 | FLJ43938 | PABP4 | iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) | Affinity Capture-MS | BioGRID | 17353931 |
| PPP1R16A | MGC14333 | MYPT3 | protein phosphatase 1, regulatory (inhibitor) subunit 16A | Two-hybrid | BioGRID | 16169070 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Two-hybrid | BioGRID | 16189514 |
| RNMTL1 | FLJ10581 | HC90 | RNA methyltransferase like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| SAP30BP | DKFZp586L2022 | HCNGP | HTRG | HTRP | SAP30 binding protein | Two-hybrid | BioGRID | 16189514 |
| SARS2 | FLJ20450 | SARS | SARSM | SERS | SYS | SerRSmt | mtSerRS | seryl-tRNA synthetase 2, mitochondrial | Affinity Capture-MS | BioGRID | 17353931 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3A3 | PRP9 | PRPF9 | SAP61 | SF3a60 | splicing factor 3a, subunit 3, 60kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS11 | DKFZp686M13204 | dJ677H15.2 | p54 | splicing factor, arginine/serine-rich 11 | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS11 | DKFZp686M13204 | dJ677H15.2 | p54 | splicing factor, arginine/serine-rich 11 | - | HPRD | 10606266 |
| SSB | LARP3 | La | Sjogren syndrome antigen B (autoantigen La) | Affinity Capture-MS | BioGRID | 17353931 |
| STOM | BND7 | EPB7 | EPB72 | stomatin | Affinity Capture-MS | BioGRID | 17353931 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| TROVE2 | RO60 | SSA2 | TROVE domain family, member 2 | Affinity Capture-MS | BioGRID | 17353931 |
| TTN | CMD1G | CMH9 | CMPD4 | CONNECTIN | DKFZp451N061 | EOMFC | FLJ26020 | FLJ26409 | FLJ32040 | FLJ34413 | FLJ39564 | FLJ43066 | HMERF | LGMD2J | TMD | titin | Affinity Capture-MS | BioGRID | 17353931 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | Affinity Capture-MS Two-hybrid | BioGRID | 16189514 |17353931 |
| UGP2 | UDPG | UDPGP2 | UGPP2 | pHC379 | UDP-glucose pyrophosphorylase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER EARLY RECURRENCE UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| OKAMOTO LIVER CANCER MULTICENTRIC OCCURRENCE UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |