Gene Page: ABCA5
Summary ?
| GeneID | 23461 |
| Symbol | ABCA5 |
| Synonyms | ABC13|EST90625 |
| Description | ATP binding cassette subfamily A member 5 |
| Reference | MIM:612503|HGNC:HGNC:35|Ensembl:ENSG00000154265|HPRD:09789|Vega:OTTHUMG00000180303 |
| Gene type | protein-coding |
| Map location | 17q24.3 |
| Pascal p-value | 0.263 |
| Sherlock p-value | 0.351 |
| Fetal beta | -1.472 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25793628 | 17 | 67311918 | ABCA5 | 3.803E-4 | 0.266 | 0.043 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9567401 | chr13 | 44860076 | ABCA5 | 23461 | 0.16 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
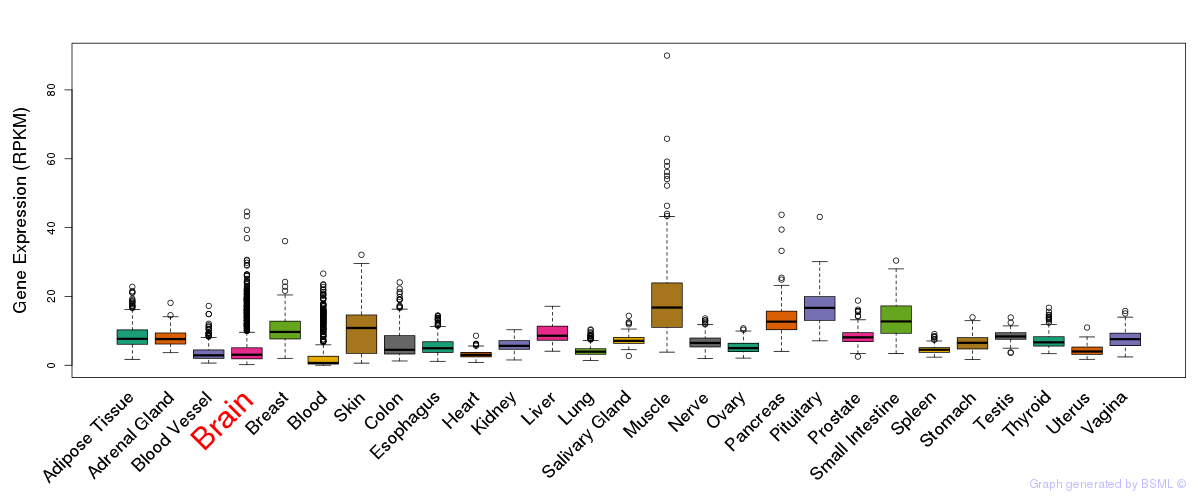
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCT6A | 0.95 | 0.96 |
| HSF2 | 0.95 | 0.95 |
| ACTR3 | 0.94 | 0.96 |
| WDR3 | 0.94 | 0.96 |
| NOL11 | 0.94 | 0.95 |
| PSPC1 | 0.94 | 0.96 |
| XRCC5 | 0.94 | 0.95 |
| DHX57 | 0.94 | 0.96 |
| POLR2B | 0.94 | 0.96 |
| PWP1 | 0.94 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.79 | -0.91 |
| AF347015.33 | -0.78 | -0.89 |
| AF347015.31 | -0.78 | -0.89 |
| HLA-F | -0.77 | -0.82 |
| FXYD1 | -0.77 | -0.89 |
| AF347015.27 | -0.76 | -0.88 |
| MT-CYB | -0.76 | -0.88 |
| AF347015.8 | -0.75 | -0.90 |
| IFI27 | -0.74 | -0.86 |
| AIFM3 | -0.74 | -0.79 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |