Gene Page: SCRIB
Summary ?
| GeneID | 23513 |
| Symbol | SCRIB |
| Synonyms | CRIB1|SCRB1|SCRIB1|Vartul |
| Description | scribbled planar cell polarity protein |
| Reference | MIM:607733|HGNC:HGNC:30377|Ensembl:ENSG00000180900|HPRD:06984|Vega:OTTHUMG00000165154 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 9.1E-6 |
| Sherlock p-value | 0.777 |
| Fetal beta | 0.455 |
| DMG | 3 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 5 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 5 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17150465 | 8 | 144897558 | SCRIB | 5.06E-5 | -0.223 | 0.022 | DMG:Wockner_2014 |
| cg23120934 | 8 | 144887083 | SCRIB | 1.858E-4 | 0.616 | 0.034 | DMG:Wockner_2014 |
| cg15650298 | 8 | 144896694 | SCRIB;MIR937 | 3.934E-4 | -0.264 | 0.043 | DMG:Wockner_2014 |
| cg08996502 | 8 | 144897297 | SCRIB | -0.029 | 0.38 | DMG:Nishioka_2013 | |
| cg02868338 | 8 | 145133133 | SCRIB | 3.661E-4 | 4.457 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
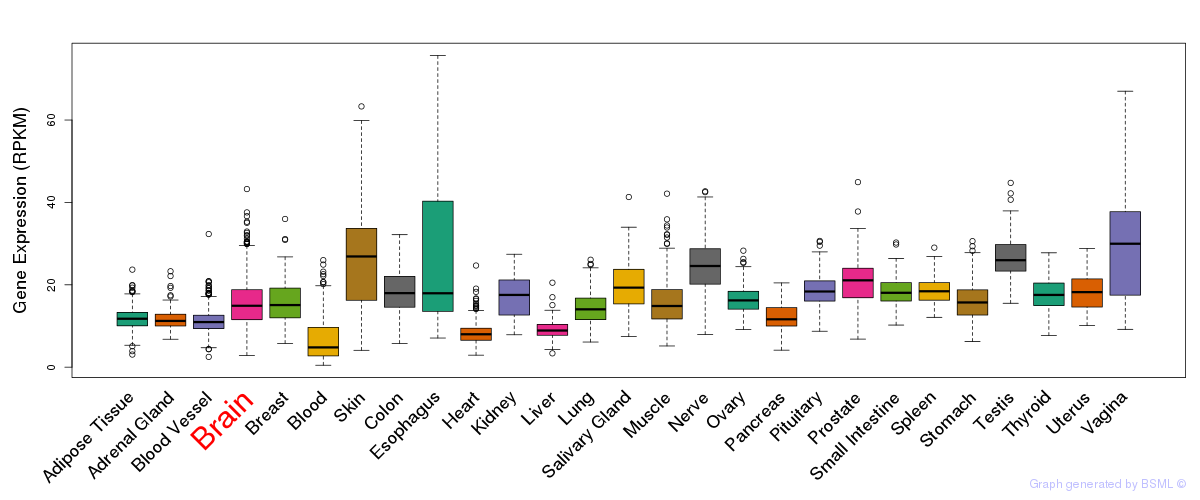
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACTR2 | 0.91 | 0.93 |
| PPP6C | 0.90 | 0.91 |
| RPRD1A | 0.90 | 0.92 |
| UCHL5 | 0.89 | 0.92 |
| PPM1A | 0.89 | 0.92 |
| G3BP2 | 0.89 | 0.91 |
| TRIM23 | 0.89 | 0.92 |
| PJA2 | 0.88 | 0.92 |
| OPA1 | 0.88 | 0.91 |
| SRPK2 | 0.87 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.61 | -0.68 |
| GSDMD | -0.61 | -0.64 |
| GATSL3 | -0.61 | -0.66 |
| FXYD1 | -0.59 | -0.57 |
| C1orf61 | -0.59 | -0.72 |
| METRN | -0.58 | -0.59 |
| CST3 | -0.58 | -0.58 |
| HIGD1B | -0.58 | -0.56 |
| S100A16 | -0.58 | -0.59 |
| EFEMP2 | -0.57 | -0.64 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GOERING BLOOD HDL CHOLESTEROL QTL TRANS | 14 | 7 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |