Gene Page: ABCA4
Summary ?
| GeneID | 24 |
| Symbol | ABCA4 |
| Synonyms | ABC10|ABCR|ARMD2|CORD3|FFM|RMP|RP19|STGD|STGD1 |
| Description | ATP binding cassette subfamily A member 4 |
| Reference | MIM:601691|HGNC:HGNC:34|Ensembl:ENSG00000198691|HPRD:03408|Vega:OTTHUMG00000010622 |
| Gene type | protein-coding |
| Map location | 1p22 |
| Pascal p-value | 0.507 |
| Fetal beta | -0.067 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09234454 | 1 | 94560697 | ABCA4 | 1E-5 | 0.777 | 0.013 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
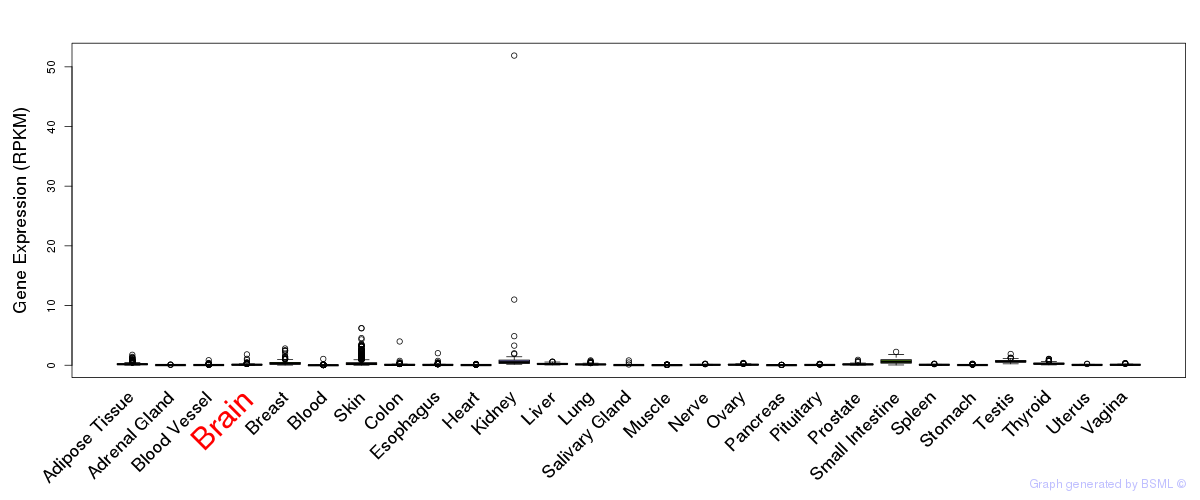
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NSD1 | 0.94 | 0.95 |
| KIF1B | 0.94 | 0.94 |
| ZZEF1 | 0.94 | 0.94 |
| NR2C2 | 0.94 | 0.94 |
| PCNX | 0.94 | 0.95 |
| MICAL3 | 0.94 | 0.95 |
| CUGBP2 | 0.93 | 0.94 |
| BIRC6 | 0.93 | 0.94 |
| TANC2 | 0.93 | 0.94 |
| KIAA0240 | 0.93 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.74 | -0.84 |
| MT-CO2 | -0.73 | -0.84 |
| AF347015.27 | -0.71 | -0.81 |
| FXYD1 | -0.71 | -0.80 |
| HIGD1B | -0.70 | -0.83 |
| AF347015.8 | -0.70 | -0.82 |
| AF347015.33 | -0.70 | -0.79 |
| AF347015.21 | -0.70 | -0.87 |
| MT-CYB | -0.69 | -0.79 |
| HSD17B14 | -0.69 | -0.76 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K27ME3 | 54 | 32 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |