Gene Page: ERC2
Summary ?
| GeneID | 26059 |
| Symbol | ERC2 |
| Synonyms | CAST|CAST1|ELKSL|SPBC110|Spc110 |
| Description | ELKS/RAB6-interacting/CAST family member 2 |
| Reference | HGNC:HGNC:31922|Ensembl:ENSG00000187672|HPRD:10810|Vega:OTTHUMG00000158390 |
| Gene type | protein-coding |
| Map location | 3p14.3 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.972 |
| Fetal beta | -0.553 |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | PROTEIN CLUSTERING G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
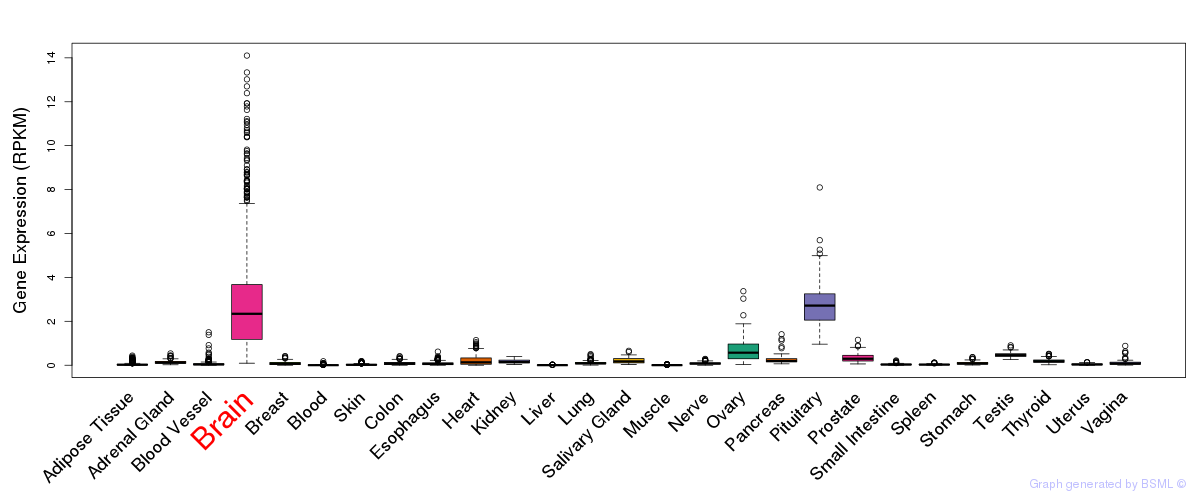
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12923177 | |
| GO:0005515 | protein binding | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042734 | presynaptic membrane | ISS | neuron, axon, Synap (GO term level: 5) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0030426 | growth cone | ISS | axon, dendrite (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 2685 | 2691 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124.1 | 1465 | 1471 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-128 | 2348 | 2355 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-135 | 1603 | 1609 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-140 | 712 | 718 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-141/200a | 2610 | 2616 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-15/16/195/424/497 | 1492 | 1498 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-182 | 725 | 731 | m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-218 | 1191 | 1197 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-24 | 1601 | 1607 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-25/32/92/363/367 | 1575 | 1581 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 2019 | 2026 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-299-5p | 711 | 717 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU | ||||
| miR-30-5p | 2336 | 2342 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-31 | 724 | 730 | 1A | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-329 | 753 | 759 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-361 | 87 | 94 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-381 | 547 | 553 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-455 | 776 | 782 | m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-485-3p | 2938 | 2944 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-490 | 685 | 691 | m8 | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-493-5p | 2910 | 2916 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-500 | 1574 | 1580 | 1A | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-9 | 13 | 19 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 2116 | 2122 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.