Gene Page: ABHD12
Summary ?
| GeneID | 26090 |
| Symbol | ABHD12 |
| Synonyms | ABHD12A|BEM46L2|C20orf22|PHARC|dJ965G21.2 |
| Description | abhydrolase domain containing 12 |
| Reference | MIM:613599|HGNC:HGNC:15868|Ensembl:ENSG00000100997|HPRD:07094|Vega:OTTHUMG00000032121 |
| Gene type | protein-coding |
| Map location | 20p11.21 |
| Pascal p-value | 0.141 |
| Sherlock p-value | 0.647 |
| Fetal beta | -1.318 |
| DMG | 1 (# studies) |
| eGene | Cortex Nucleus accumbens basal ganglia Myers' cis & trans Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Expression | Meta-analysis of gene expression | P value: 1.631 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABHD12 | chr20 | 25290190 | G | A | NM_001042472 NM_015600 | p.214T>M p.214T>M | missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17351116 | 20 | 25371719 | ABHD12 | 3.67E-10 | -0.01 | 7.44E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6138506 | chr20 | 25115635 | ABHD12 | 26090 | 0.18 | cis | ||
| rs2268879 | chr20 | 25177804 | ABHD12 | 26090 | 0.01 | cis | ||
| rs6050422 | chr20 | 25177919 | ABHD12 | 26090 | 0.01 | cis | ||
| rs6083776 | chr20 | 25186115 | ABHD12 | 26090 | 5.564E-7 | cis | ||
| rs7453 | chr20 | 25207073 | ABHD12 | 26090 | 2.349E-16 | cis | ||
| rs3746337 | chr20 | 25208271 | ABHD12 | 26090 | 5.858E-11 | cis | ||
| rs6132819 | chr20 | 25218617 | ABHD12 | 26090 | 3.314E-16 | cis | ||
| rs8184820 | chr20 | 25250576 | ABHD12 | 26090 | 1.026E-9 | cis | ||
| rs2257233 | chr20 | 25265983 | ABHD12 | 26090 | 2.687E-18 | cis | ||
| rs2257991 | chr20 | 25270338 | ABHD12 | 26090 | 2.007E-18 | cis | ||
| rs2474777 | chr20 | 25272632 | ABHD12 | 26090 | 7.986E-18 | cis | ||
| rs2258719 | chr20 | 25275842 | ABHD12 | 26090 | 7.986E-18 | cis | ||
| rs1888999 | chr20 | 25291847 | ABHD12 | 26090 | 7.986E-18 | cis | ||
| rs2297497 | chr20 | 25297908 | ABHD12 | 26090 | 2.687E-18 | cis | ||
| rs7267979 | chr20 | 25298086 | ABHD12 | 26090 | 7.986E-18 | cis | ||
| rs2274890 | chr20 | 25301021 | ABHD12 | 26090 | 3.82E-8 | cis | ||
| rs6050559 | chr20 | 25329914 | ABHD12 | 26090 | 1.914E-17 | cis | ||
| rs6050565 | chr20 | 25347562 | ABHD12 | 26090 | 5.359E-16 | cis | ||
| rs4815411 | chr20 | 25350222 | ABHD12 | 26090 | 1.025E-18 | cis | ||
| rs4815412 | chr20 | 25350324 | ABHD12 | 26090 | 1.025E-18 | cis | ||
| rs6050573 | chr20 | 25366064 | ABHD12 | 26090 | 1.374E-18 | cis | ||
| rs6050598 | chr20 | 25397256 | ABHD12 | 26090 | 1.025E-18 | cis | ||
| rs12428 | chr20 | 25433820 | ABHD12 | 26090 | 7.882E-19 | cis | ||
| rs449370 | chr20 | 25451179 | ABHD12 | 26090 | 1.99E-18 | cis | ||
| rs376742 | chr20 | 25487416 | ABHD12 | 26090 | 5.987E-17 | cis | ||
| rs7272126 | chr20 | 25488883 | ABHD12 | 26090 | 0.03 | cis | ||
| rs16987806 | chr20 | 25492189 | ABHD12 | 26090 | 0.03 | cis | ||
| rs6115202 | chr20 | 25497537 | ABHD12 | 26090 | 1.177E-7 | cis | ||
| rs443009 | chr20 | 25499816 | ABHD12 | 26090 | 5.538E-16 | cis | ||
| rs6138598 | chr20 | 25513535 | ABHD12 | 26090 | 0.06 | cis | ||
| rs6083877 | chr20 | 25520938 | ABHD12 | 26090 | 3.218E-4 | cis | ||
| rs1983974 | chr20 | 25530618 | ABHD12 | 26090 | 5.987E-17 | cis | ||
| rs6138601 | chr20 | 25539485 | ABHD12 | 26090 | 0.11 | cis | ||
| rs6132862 | chr20 | 25721247 | ABHD12 | 26090 | 0.15 | cis | ||
| rs6115458 | chr20 | 25969264 | ABHD12 | 26090 | 6.475E-6 | cis | ||
| rs845778 | chr20 | 26167546 | ABHD12 | 26090 | 0.03 | cis | ||
| rs845779 | chr20 | 26167979 | ABHD12 | 26090 | 9.858E-6 | cis | ||
| rs845794 | chr20 | 26222698 | ABHD12 | 26090 | 0.06 | cis | ||
| rs845796 | chr20 | 26225144 | ABHD12 | 26090 | 0.16 | cis | ||
| rs6083776 | chr20 | 25186115 | ABHD12 | 26090 | 1.094E-4 | trans | ||
| rs7453 | chr20 | 25207073 | ABHD12 | 26090 | 5.535E-14 | trans | ||
| rs3746337 | chr20 | 25208271 | ABHD12 | 26090 | 1.507E-8 | trans | ||
| rs6132819 | chr20 | 25218617 | ABHD12 | 26090 | 7.891E-14 | trans | ||
| rs8183079 | 0 | ABHD12 | 26090 | 9.312E-9 | trans | |||
| rs8184820 | chr20 | 25250576 | ABHD12 | 26090 | 2.533E-7 | trans | ||
| rs2257233 | chr20 | 25265983 | ABHD12 | 26090 | 5.421E-16 | trans | ||
| rs2257991 | chr20 | 25270338 | ABHD12 | 26090 | 3.905E-16 | trans | ||
| rs2474777 | chr20 | 25272632 | ABHD12 | 26090 | 1.778E-15 | trans | ||
| rs2258719 | chr20 | 25275842 | ABHD12 | 26090 | 1.778E-15 | trans | ||
| rs1888999 | chr20 | 25291847 | ABHD12 | 26090 | 1.778E-15 | trans | ||
| rs2297497 | chr20 | 25297908 | ABHD12 | 26090 | 5.443E-16 | trans | ||
| rs7267979 | chr20 | 25298086 | ABHD12 | 26090 | 1.818E-15 | trans | ||
| rs2274890 | chr20 | 25301021 | ABHD12 | 26090 | 8.87E-6 | trans | ||
| rs6050559 | chr20 | 25329914 | ABHD12 | 26090 | 4.451E-15 | trans | ||
| rs6050565 | chr20 | 25347562 | ABHD12 | 26090 | 1.313E-13 | trans | ||
| rs4815411 | chr20 | 25350222 | ABHD12 | 26090 | 1.71E-16 | trans | ||
| rs4815412 | chr20 | 25350324 | ABHD12 | 26090 | 1.71E-16 | trans | ||
| rs6050573 | chr20 | 25366064 | ABHD12 | 26090 | 2.309E-16 | trans | ||
| rs6515641 | 0 | ABHD12 | 26090 | 5.802E-11 | trans | |||
| rs6050598 | chr20 | 25397256 | ABHD12 | 26090 | 1.699E-16 | trans | ||
| rs6417589 | 0 | ABHD12 | 26090 | 1.317E-13 | trans | |||
| rs12428 | chr20 | 25433820 | ABHD12 | 26090 | 1.151E-16 | trans | ||
| rs449370 | chr20 | 25451179 | ABHD12 | 26090 | 3.524E-16 | trans | ||
| rs376742 | chr20 | 25487416 | ABHD12 | 26090 | 1.448E-14 | trans | ||
| rs6115202 | chr20 | 25497537 | ABHD12 | 26090 | 2.566E-5 | trans | ||
| rs443009 | chr20 | 25499816 | ABHD12 | 26090 | 1.379E-13 | trans | ||
| rs6083877 | chr20 | 25520938 | ABHD12 | 26090 | 0.03 | trans | ||
| rs1983974 | chr20 | 25530618 | ABHD12 | 26090 | 1.448E-14 | trans | ||
| rs6115458 | chr20 | 25969264 | ABHD12 | 26090 | 0 | trans | ||
| rs845779 | chr20 | 26167979 | ABHD12 | 26090 | 0 | trans |
Section II. Transcriptome annotation
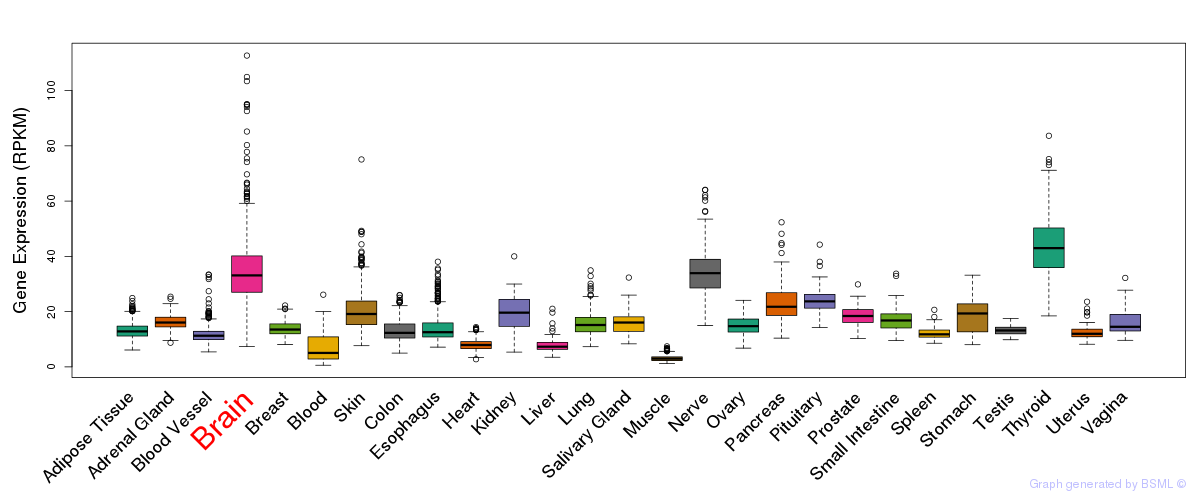
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY UP | 109 | 69 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MF UP | 47 | 27 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |