Gene Page: GGCX
Summary ?
| GeneID | 2677 |
| Symbol | GGCX |
| Synonyms | VKCFD1 |
| Description | gamma-glutamyl carboxylase |
| Reference | MIM:137167|HGNC:HGNC:4247|Ensembl:ENSG00000115486|HPRD:00665|Vega:OTTHUMG00000130173 |
| Gene type | protein-coding |
| Map location | 2p12 |
| Pascal p-value | 0.265 |
| Sherlock p-value | 0.04 |
| Fetal beta | -0.449 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hypothalamus Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13174083 | 2 | 85788758 | GGCX | 2.73E-9 | -0.025 | 1.95E-6 | DMG:Jaffe_2016 |
| cg17127132 | 2 | 85788382 | GGCX | 5.31E-8 | -0.016 | 1.38E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6738009 | 2 | 85737509 | GGCX | ENSG00000115486.7 | 1.759E-7 | 0 | 51161 | gtex_brain_ba24 |
| rs7591175 | 2 | 85738374 | GGCX | ENSG00000115486.7 | 6.655E-7 | 0 | 50296 | gtex_brain_ba24 |
| rs2166529 | 2 | 85742175 | GGCX | ENSG00000115486.7 | 1.3E-7 | 0 | 46495 | gtex_brain_ba24 |
| rs6750847 | 2 | 85758144 | GGCX | ENSG00000115486.7 | 1.458E-6 | 0 | 30526 | gtex_brain_ba24 |
| rs6731005 | 2 | 85760395 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | 28275 | gtex_brain_ba24 |
| rs6705839 | 2 | 85761279 | GGCX | ENSG00000115486.7 | 1.188E-7 | 0 | 27391 | gtex_brain_ba24 |
| rs6705971 | 2 | 85761417 | GGCX | ENSG00000115486.7 | 2.759E-7 | 0 | 27253 | gtex_brain_ba24 |
| rs10175792 | 2 | 85761654 | GGCX | ENSG00000115486.7 | 2.409E-7 | 0 | 27016 | gtex_brain_ba24 |
| rs10176176 | 2 | 85762048 | GGCX | ENSG00000115486.7 | 5.893E-8 | 0 | 26622 | gtex_brain_ba24 |
| rs10179195 | 2 | 85762761 | GGCX | ENSG00000115486.7 | 2.749E-7 | 0 | 25909 | gtex_brain_ba24 |
| rs6714157 | 2 | 85763274 | GGCX | ENSG00000115486.7 | 2.028E-6 | 0 | 25396 | gtex_brain_ba24 |
| rs6743030 | 2 | 85763520 | GGCX | ENSG00000115486.7 | 2.744E-7 | 0 | 25150 | gtex_brain_ba24 |
| rs3755014 | 2 | 85764006 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | 24664 | gtex_brain_ba24 |
| rs3755015 | 2 | 85764041 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | 24629 | gtex_brain_ba24 |
| rs1446668 | 2 | 85764960 | GGCX | ENSG00000115486.7 | 9.1E-7 | 0 | 23710 | gtex_brain_ba24 |
| rs2028900 | 2 | 85767735 | GGCX | ENSG00000115486.7 | 1.842E-7 | 0 | 20935 | gtex_brain_ba24 |
| rs1078004 | 2 | 85769711 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | 18959 | gtex_brain_ba24 |
| rs7605975 | 2 | 85772548 | GGCX | ENSG00000115486.7 | 5.888E-8 | 0 | 16122 | gtex_brain_ba24 |
| rs12473819 | 2 | 85773061 | GGCX | ENSG00000115486.7 | 2.064E-7 | 0 | 15609 | gtex_brain_ba24 |
| rs6547621 | 2 | 85774676 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | 13994 | gtex_brain_ba24 |
| rs5832649 | 2 | 85776490 | GGCX | ENSG00000115486.7 | 7.418E-8 | 0 | 12180 | gtex_brain_ba24 |
| rs6738645 | 2 | 85783128 | GGCX | ENSG00000115486.7 | 5.881E-8 | 0 | 5542 | gtex_brain_ba24 |
| rs7568458 | 2 | 85788175 | GGCX | ENSG00000115486.7 | 7.041E-8 | 0 | 495 | gtex_brain_ba24 |
| rs11891260 | 2 | 85796101 | GGCX | ENSG00000115486.7 | 2.064E-7 | 0 | -7431 | gtex_brain_ba24 |
| rs1561198 | 2 | 85809989 | GGCX | ENSG00000115486.7 | 5.769E-8 | 0 | -21319 | gtex_brain_ba24 |
| rs55971080 | 2 | 85812746 | GGCX | ENSG00000115486.7 | 1.745E-8 | 0 | -24076 | gtex_brain_ba24 |
Section II. Transcriptome annotation
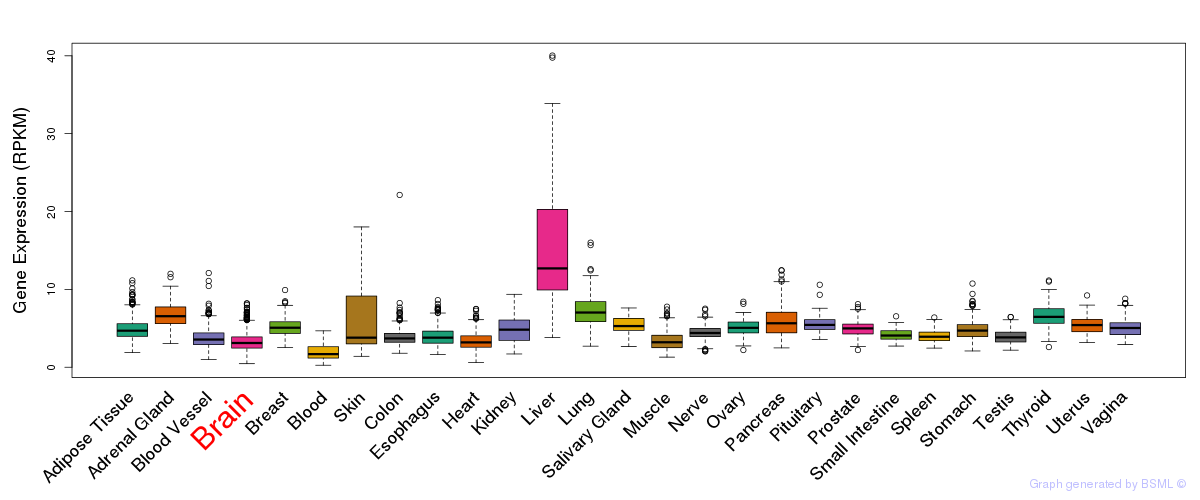
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008488 | gamma-glutamyl carboxylase activity | TAS | 1749935 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006464 | protein modification process | TAS | 1749935 | |
| GO:0007596 | blood coagulation | TAS | 9845520 | |
| GO:0017187 | peptidyl-glutamic acid carboxylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 1749935 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | 1749935 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |