Gene Page: CHIA
Summary ?
| GeneID | 27159 |
| Symbol | CHIA |
| Synonyms | AMCASE|CHIT2|TSA1902 |
| Description | chitinase, acidic |
| Reference | MIM:606080|HGNC:HGNC:17432|Ensembl:ENSG00000134216|HPRD:09355|Vega:OTTHUMG00000011165 |
| Gene type | protein-coding |
| Map location | 1p13.2 |
| Pascal p-value | 0.411 |
| Fetal beta | -0.283 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
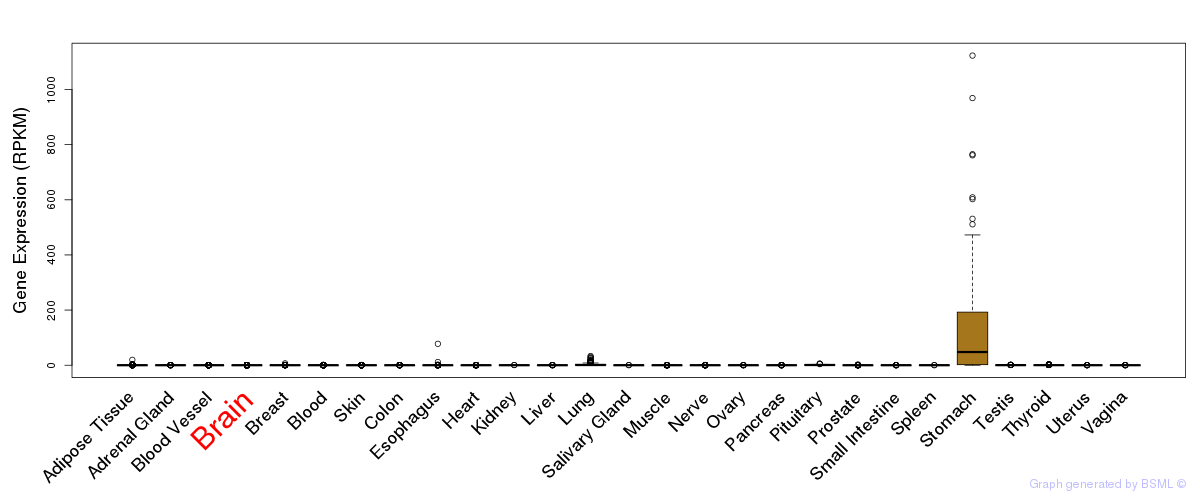
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0003796 | lysozyme activity | NAS | 11085997 | |
| GO:0005529 | sugar binding | NAS | 11085997 | |
| GO:0004568 | chitinase activity | IDA | 11085997 | |
| GO:0004568 | chitinase activity | IEA | - | |
| GO:0004568 | chitinase activity | NAS | 10548734 | |
| GO:0008061 | chitin binding | IEA | - | |
| GO:0008061 | chitin binding | TAS | 11085997 | |
| GO:0043169 | cation binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000272 | polysaccharide catabolic process | IEA | - | |
| GO:0001101 | response to acid | TAS | 11085997 | |
| GO:0006037 | cell wall chitin metabolic process | TAS | 11085997 | |
| GO:0006032 | chitin catabolic process | IEA | - | |
| GO:0006032 | chitin catabolic process | NAS | 11085997 | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0009620 | response to fungus | TAS | 11085997 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007586 | digestion | NAS | 11085997 | |
| GO:0006955 | immune response | NAS | 11085997 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IC | 11085997 | |
| GO:0005737 | cytoplasm | NAS | 10548734 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| MAEKAWA ATF2 TARGETS | 24 | 19 | All SZGR 2.0 genes in this pathway |