Gene Page: GLI3
Summary ?
| GeneID | 2737 |
| Symbol | GLI3 |
| Synonyms | ACLS|GCPS|GLI3-190|GLI3FL|PAP-A|PAPA|PAPA1|PAPB|PHS|PPDIV |
| Description | GLI family zinc finger 3 |
| Reference | MIM:165240|HGNC:HGNC:4319|Ensembl:ENSG00000106571|HPRD:01313|Vega:OTTHUMG00000023630 |
| Gene type | protein-coding |
| Map location | 7p13 |
| Pascal p-value | 0.053 |
| Sherlock p-value | 0.414 |
| TADA p-value | 0.028 |
| Fetal beta | 3.141 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLI3 | chr7 | 42079692 | G | A | NM_000168 | p.325R>C | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs846300 | chr7 | 42055260 | GLI3 | 2737 | 0.06 | cis | ||
| rs17301762 | chr1 | 180794652 | GLI3 | 2737 | 0.07 | trans | ||
| rs17302102 | chr1 | 180866344 | GLI3 | 2737 | 0.07 | trans | ||
| rs17801458 | chr2 | 142468471 | GLI3 | 2737 | 0.02 | trans | ||
| rs16870771 | chr4 | 21307780 | GLI3 | 2737 | 0.05 | trans | ||
| rs6879593 | chr5 | 75253310 | GLI3 | 2737 | 0.03 | trans | ||
| rs17061935 | chr6 | 101873751 | GLI3 | 2737 | 0.06 | trans | ||
| rs2225177 | chr10 | 108777243 | GLI3 | 2737 | 0.14 | trans | ||
| rs1111195 | chr16 | 51416325 | GLI3 | 2737 | 0.15 | trans | ||
| rs9944345 | chr16 | 51419164 | GLI3 | 2737 | 0.15 | trans | ||
| rs16981461 | chr20 | 56490699 | GLI3 | 2737 | 0.13 | trans | ||
| rs3099706 | 0 | GLI3 | 2737 | 0.16 | trans |
Section II. Transcriptome annotation
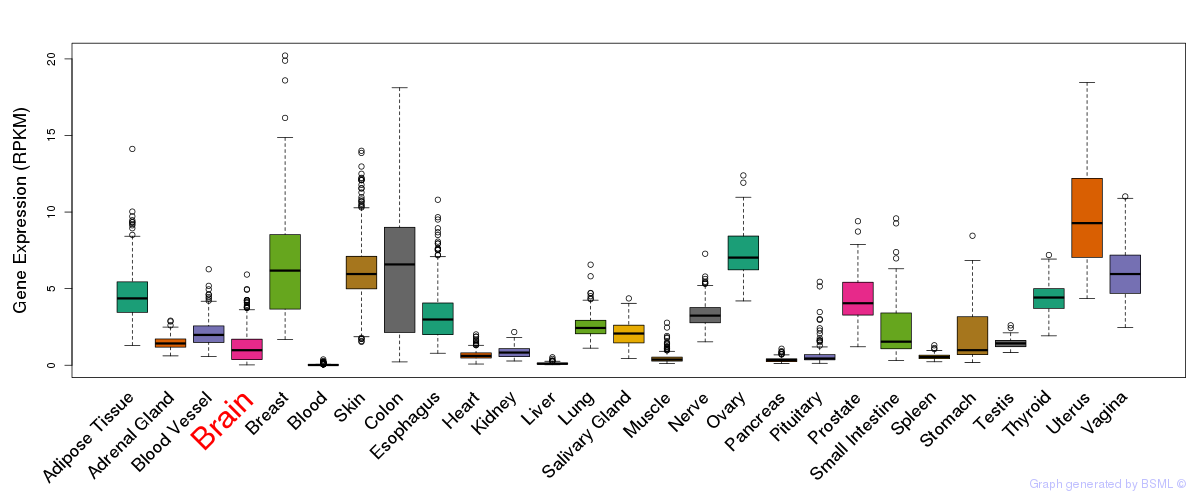
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 9054938 |10077605 | |
| GO:0005515 | protein binding | IPI | 10564661 |10806483 |11238441 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048663 | neuron fate commitment | IEA | neuron (GO term level: 9) | - |
| GO:0021631 | optic nerve morphogenesis | IEA | Brain (GO term level: 9) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0021915 | neural tube development | IEA | Brain (GO term level: 7) | - |
| GO:0021776 | smoothened signaling pathway involved in spinal cord motor neuron cell fate specification | IEA | neuron (GO term level: 12) | - |
| GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification | IEA | neuron (GO term level: 12) | - |
| GO:0000060 | protein import into nucleus, translocation | TAS | 10077605 | |
| GO:0001658 | ureteric bud branching | IEA | - | |
| GO:0001656 | metanephros development | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009954 | proximal/distal pattern formation | IEA | - | |
| GO:0007224 | smoothened signaling pathway | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10077605 | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0007442 | hindgut morphogenesis | IEA | - | |
| GO:0007389 | pattern specification process | IEA | - | |
| GO:0016481 | negative regulation of transcription | IEA | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0030879 | mammary gland development | IEA | - | |
| GO:0030326 | embryonic limb morphogenesis | IEA | - | |
| GO:0030324 | lung development | IEA | - | |
| GO:0035295 | tube development | IEA | - | |
| GO:0021513 | spinal cord dorsal/ventral patterning | IEA | - | |
| GO:0045596 | negative regulation of cell differentiation | IEA | - | |
| GO:0048558 | embryonic gut morphogenesis | IEA | - | |
| GO:0048589 | developmental growth | IEA | - | |
| GO:0048593 | camera-type eye morphogenesis | IEA | - | |
| GO:0048598 | embryonic morphogenesis | IEA | - | |
| GO:0048646 | anatomical structure formation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | TAS | 10077605 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | TAS | 10077605 | |
| GO:0017053 | transcriptional repressor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 10075717 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD | 9843199 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 9843199 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD | 9843199 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD | 9843199 |
| STK36 | DKFZp434N0223 | FU | KIAA1278 | serine/threonine kinase 36, fused homolog (Drosophila) | - | HPRD,BioGRID | 10806483 |
| SUFU | PRO1280 | SUFUH | SUFUXL | suppressor of fused homolog (Drosophila) | - | HPRD,BioGRID | 10564661 |
| TWIST1 | ACS3 | BPES2 | BPES3 | SCS | TWIST | bHLHa38 | twist homolog 1 (Drosophila) | - | HPRD | 12142027 |
| ZIC1 | ZIC | ZNF201 | Zic family member 1 (odd-paired homolog, Drosophila) | - | HPRD,BioGRID | 11238441 |
| ZIC2 | HPE5 | Zic family member 2 (odd-paired homolog, Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11238441 |
| ZIC3 | HTX | HTX1 | ZNF203 | Zic family member 3 (odd-paired homolog, Drosophila) | Reconstituted Complex | BioGRID | 11238441 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP DN | 35 | 20 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 DN | 44 | 28 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR | 27 | 12 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 2859 | 2865 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 2858 | 2865 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-143 | 107 | 114 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-200bc/429 | 217 | 223 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 201 | 207 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-24 | 2826 | 2832 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-494 | 3333 | 3339 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-496 | 3149 | 3156 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-7 | 195 | 202 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.