Gene Page: GPR6
Summary ?
| GeneID | 2830 |
| Symbol | GPR6 |
| Synonyms | - |
| Description | G protein-coupled receptor 6 |
| Reference | MIM:600553|HGNC:HGNC:4515|Ensembl:ENSG00000146360|HPRD:02775| |
| Gene type | protein-coding |
| Map location | 6q21 |
| Pascal p-value | 0.222 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6924963 | chr6 | 110838907 | GPR6 | 2830 | 0.01 | cis | ||
| rs9386913 | chr6 | 110850514 | GPR6 | 2830 | 0.12 | cis | ||
| rs17186508 | chr5 | 65194022 | GPR6 | 2830 | 0.18 | trans | ||
| rs1329778 | chr9 | 73755912 | GPR6 | 2830 | 0.17 | trans | ||
| rs16976379 | chr18 | 40198836 | GPR6 | 2830 | 0.04 | trans |
Section II. Transcriptome annotation
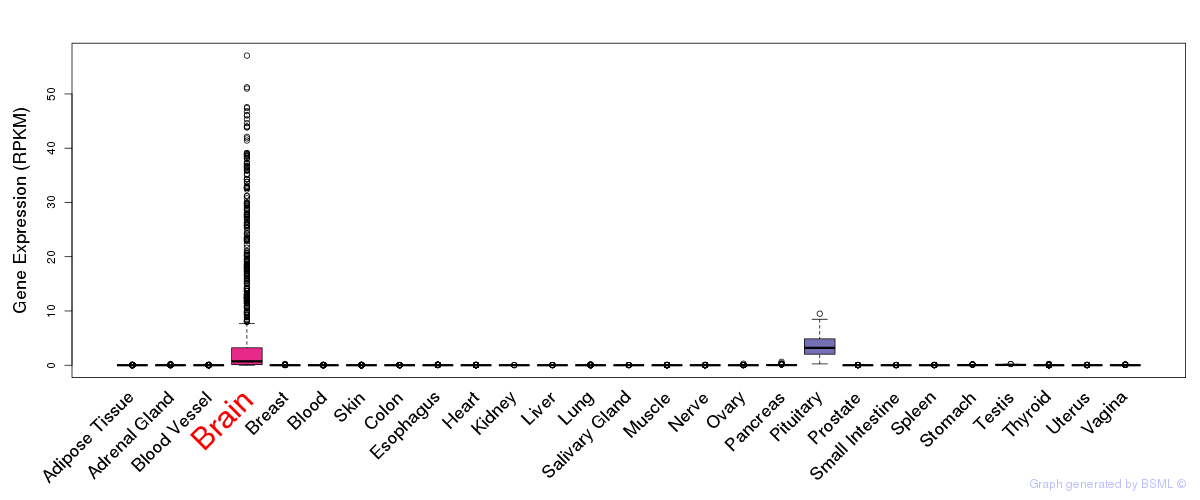
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TOLLIP | 0.89 | 0.87 |
| KCNK1 | 0.89 | 0.82 |
| DNM1 | 0.89 | 0.88 |
| EXTL1 | 0.89 | 0.84 |
| CNTNAP1 | 0.89 | 0.89 |
| FAIM2 | 0.89 | 0.90 |
| MFSD4 | 0.89 | 0.88 |
| TMEM130 | 0.88 | 0.89 |
| TYRO3 | 0.88 | 0.82 |
| MAST3 | 0.88 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTF3C6 | -0.50 | -0.47 |
| PDE9A | -0.49 | -0.53 |
| BCL7C | -0.48 | -0.55 |
| RBMX2 | -0.48 | -0.56 |
| C9orf46 | -0.48 | -0.51 |
| FAM36A | -0.48 | -0.39 |
| FADS2 | -0.47 | -0.42 |
| TRAF4 | -0.47 | -0.53 |
| TUBB2B | -0.47 | -0.52 |
| KIAA1949 | -0.46 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001619 | lysosphingolipid and lysophosphatidic acid receptor activity | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007204 | elevation of cytosolic calcium ion concentration | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 7832990 | |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7832990 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER H3K27ME3 IN NORMAL AND METHYLATED IN CANCER | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |