Gene Page: GPS1
Summary ?
| GeneID | 2873 |
| Symbol | GPS1 |
| Synonyms | COPS1|CSN1|SGN1 |
| Description | G protein pathway suppressor 1 |
| Reference | MIM:601934|HGNC:HGNC:4549|Ensembl:ENSG00000169727|HPRD:15991|Vega:OTTHUMG00000178484 |
| Gene type | protein-coding |
| Map location | 17q25.3 |
| Pascal p-value | 0.041 |
| Sherlock p-value | 0.299 |
| Fetal beta | -0.145 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24392479 | 17 | 79934870 | GPS1 | 4.528E-4 | 5.6 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
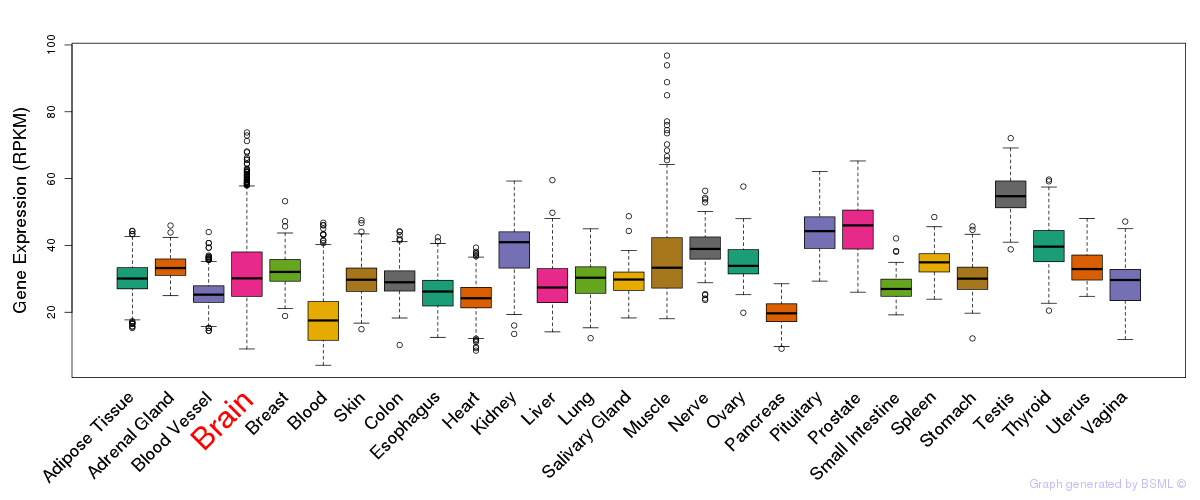
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDXDC1 | 0.82 | 0.89 |
| PTPN1 | 0.82 | 0.91 |
| GRB2 | 0.82 | 0.88 |
| HEATR5B | 0.81 | 0.88 |
| SPIN3 | 0.81 | 0.88 |
| ZMYND11 | 0.81 | 0.89 |
| BAI3 | 0.81 | 0.87 |
| C4orf41 | 0.81 | 0.87 |
| STX6 | 0.81 | 0.88 |
| B4GALT5 | 0.81 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.81 |
| MT-CO2 | -0.70 | -0.81 |
| HIGD1B | -0.69 | -0.82 |
| FXYD1 | -0.69 | -0.78 |
| AF347015.33 | -0.69 | -0.77 |
| MT-CYB | -0.67 | -0.77 |
| AF347015.27 | -0.67 | -0.77 |
| AF347015.8 | -0.66 | -0.78 |
| AF347015.2 | -0.66 | -0.78 |
| AC021016.1 | -0.66 | -0.77 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | CSN1 interacts with CSN2. | BIND | 12615944 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD | 11114242 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | - | HPRD | 11114242 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | CSN1 interacts with CSN4. | BIND | 12615944 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | - | HPRD | 11114242 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | Co-purification | BioGRID | 9707402 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | - | HPRD | 11337588 |
| CUL1 | MGC149834 | MGC149835 | cullin 1 | - | HPRD | 11337588 |
| CUL2 | MGC131970 | cullin 2 | - | HPRD | 11337588 |
| CUL3 | - | cullin 3 | - | HPRD | 11337588 |
| ITPK1 | ITRPK1 | inositol 1,3,4-triphosphate 5/6 kinase | - | HPRD | 12324474 |
| RBX1 | BA554C12.1 | MGC13357 | MGC1481 | RNF75 | ROC1 | ring-box 1 | - | HPRD | 11337588 |
| SKP1 | EMC19 | MGC34403 | OCP-II | OCP2 | SKP1A | TCEB1L | p19A | S-phase kinase-associated protein 1 | - | HPRD | 11337588 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | - | HPRD | 11337588 |
| WDR8 | FLJ20430 | MGC99569 | WD repeat domain 8 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS UP | 40 | 20 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |