Gene Page: GTF2E2
Summary ?
| GeneID | 2961 |
| Symbol | GTF2E2 |
| Synonyms | FE|TF2E2|TFIIE-B |
| Description | general transcription factor IIE subunit 2 |
| Reference | MIM:189964|HGNC:HGNC:4651|Ensembl:ENSG00000197265|HPRD:01800|Vega:OTTHUMG00000163934 |
| Gene type | protein-coding |
| Map location | 8p12 |
| Pascal p-value | 0.331 |
| Sherlock p-value | 0.121 |
| Fetal beta | 0.535 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18356330 | 8 | 30515990 | GTF2E2 | -0.021 | 0.28 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs423974 | chr16 | 77930523 | GTF2E2 | 2961 | 0.13 | trans |
Section II. Transcriptome annotation
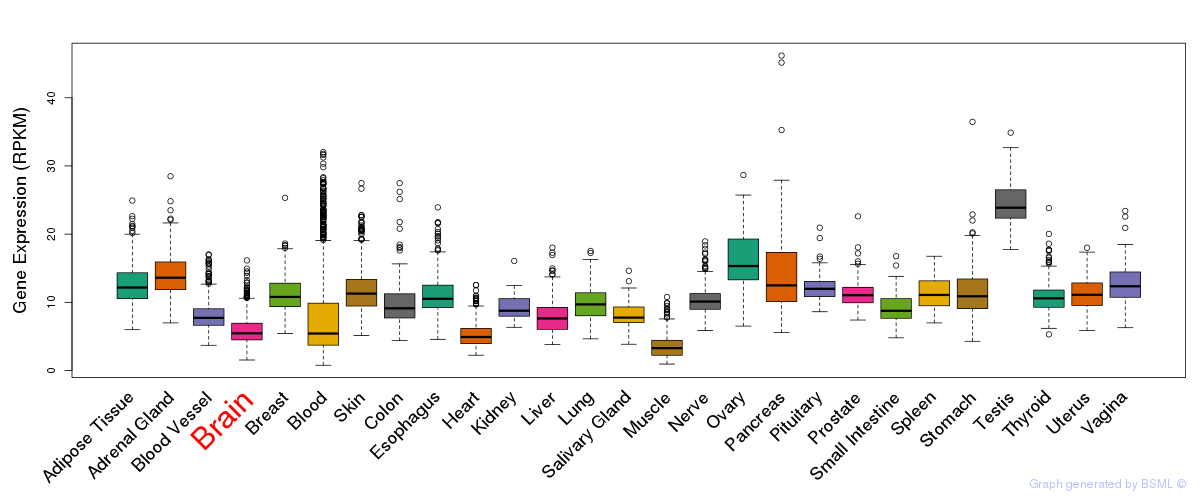
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMARCD3 | 0.74 | 0.75 |
| CDKN2D | 0.74 | 0.72 |
| ETNK2 | 0.74 | 0.76 |
| FKBP1B | 0.74 | 0.76 |
| RNF7 | 0.72 | 0.74 |
| VPS25 | 0.71 | 0.71 |
| CHID1 | 0.70 | 0.73 |
| C1orf59 | 0.69 | 0.71 |
| CHCHD6 | 0.68 | 0.71 |
| GNB2 | 0.68 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.52 | -0.56 |
| MT-CO2 | -0.50 | -0.53 |
| AF347015.8 | -0.50 | -0.54 |
| AF347015.26 | -0.50 | -0.56 |
| AF347015.15 | -0.50 | -0.54 |
| AF347015.33 | -0.49 | -0.54 |
| MT-CYB | -0.49 | -0.54 |
| AF347015.27 | -0.48 | -0.54 |
| AF347015.31 | -0.47 | -0.50 |
| AF347015.9 | -0.46 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003702 | RNA polymerase II transcription factor activity | IEA | - | |
| GO:0005515 | protein binding | IDA | 9271120 | |
| GO:0005515 | protein binding | IPI | 7926747 |8999876 |9271120 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | EXP | 8946909 | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | TAS | 1956398 | |
| GO:0006368 | RNA elongation from RNA polymerase II promoter | EXP | 9405375 | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 1939271 |2449431 |8946909 | |
| GO:0005673 | transcription factor TFIIE complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATF7IP | FLJ10139 | FLJ10688 | MCAF | p621 | activating transcription factor 7 interacting protein | - | HPRD | 10777215 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | Reconstituted Complex | BioGRID | 7926747 |
| ERCC6 | ARMD5 | CKN2 | COFS | COFS1 | CSB | RAD26 | excision repair cross-complementing rodent repair deficiency, complementation group 6 | - | HPRD | 8999876 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | Reconstituted Complex | BioGRID | 8628277 |
| GTF2A1 | MGC129969 | MGC129970 | TF2A1 | TFIIA | general transcription factor IIA, 1, 19/37kDa | - | HPRD,BioGRID | 9618479 |11509574 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | Affinity Capture-Western Reconstituted Complex | BioGRID | 11113176 |12665589 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | - | HPRD,BioGRID | 9677423 |11113176 |12665589 |
| GTF2E2 | FE | TF2E2 | TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa | Reconstituted Complex | BioGRID | 9677423 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Reconstituted Complex | BioGRID | 9677423 |11113176 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | Reconstituted Complex | BioGRID | 11113176 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | Reconstituted Complex | BioGRID | 8628277 |
| KLF5 | BTEB2 | CKLF | IKLF | Kruppel-like factor 5 (intestinal) | - | HPRD | 9089417 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 9271120 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western | BioGRID | 12665589 |
| SND1 | TDRD11 | p100 | staphylococcal nuclease and tudor domain containing 1 | - | HPRD,BioGRID | 7651391 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 9677423 |11113176 |
| XPA | XP1 | XPAC | xeroderma pigmentosum, complementation group A | Reconstituted Complex | BioGRID | 7876263 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASAL TRANSCRIPTION FACTORS | 36 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| SILIGAN TARGETS OF EWS FLI1 FUSION DN | 18 | 9 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE UP | 50 | 36 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-30-5p | 121 | 127 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.