Gene Page: FTSJ2
Summary ?
| GeneID | 29960 |
| Symbol | FTSJ2 |
| Synonyms | FJH1|HEL97|MRM2|RRMJ2 |
| Description | FtsJ RNA methyltransferase homolog 2 (E. coli) |
| Reference | MIM:606906|HGNC:HGNC:16352|Ensembl:ENSG00000122687|HPRD:06067|Vega:OTTHUMG00000023866 |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 2.245E-5 |
| Fetal beta | -0.032 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs71525363 | 7 | 2268792 | FTSJ2 | ENSG00000122687.13 | 2.41E-6 | 0.02 | 13048 | gtex_brain_putamen_basal |
| rs3757437 | 7 | 2271980 | FTSJ2 | ENSG00000122687.13 | 2.352E-6 | 0.02 | 9860 | gtex_brain_putamen_basal |
| rs34418140 | 7 | 2278183 | FTSJ2 | ENSG00000122687.13 | 2.352E-6 | 0.02 | 3657 | gtex_brain_putamen_basal |
| rs2286207 | 7 | 2303109 | FTSJ2 | ENSG00000122687.13 | 1.705E-6 | 0.02 | -21269 | gtex_brain_putamen_basal |
| rs34219261 | 7 | 2326816 | FTSJ2 | ENSG00000122687.13 | 1.769E-6 | 0.02 | -44976 | gtex_brain_putamen_basal |
| rs398003402 | 7 | 2326881 | FTSJ2 | ENSG00000122687.13 | 1.752E-6 | 0.02 | -45041 | gtex_brain_putamen_basal |
| rs4721548 | 7 | 2330496 | FTSJ2 | ENSG00000122687.13 | 2.042E-6 | 0.02 | -48656 | gtex_brain_putamen_basal |
| rs75537891 | 7 | 2332880 | FTSJ2 | ENSG00000122687.13 | 1.105E-6 | 0.02 | -51040 | gtex_brain_putamen_basal |
| rs12699820 | 7 | 2332968 | FTSJ2 | ENSG00000122687.13 | 1.074E-6 | 0.02 | -51128 | gtex_brain_putamen_basal |
| rs34974704 | 7 | 2333057 | FTSJ2 | ENSG00000122687.13 | 1.065E-6 | 0.02 | -51217 | gtex_brain_putamen_basal |
| rs7797577 | 7 | 2333542 | FTSJ2 | ENSG00000122687.13 | 1.585E-6 | 0.02 | -51702 | gtex_brain_putamen_basal |
| rs13233723 | 7 | 2338087 | FTSJ2 | ENSG00000122687.13 | 7.992E-7 | 0.02 | -56247 | gtex_brain_putamen_basal |
| rs7781284 | 7 | 2339405 | FTSJ2 | ENSG00000122687.13 | 1.834E-6 | 0.02 | -57565 | gtex_brain_putamen_basal |
| rs4721561 | 7 | 2340117 | FTSJ2 | ENSG00000122687.13 | 1.547E-6 | 0.02 | -58277 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
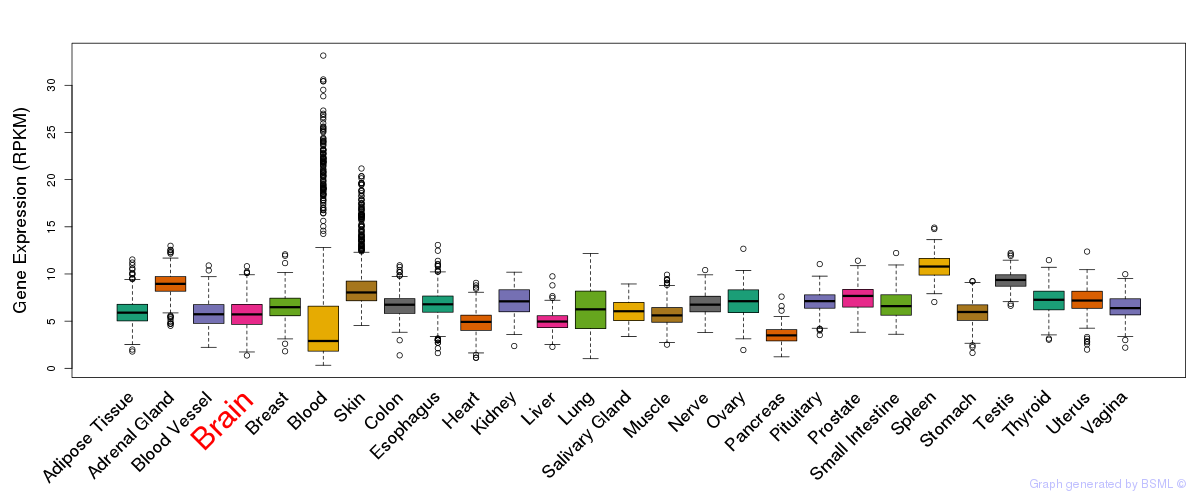
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 MCF10A | 43 | 26 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |