Gene Page: HLA-DOB
Summary ?
| GeneID | 3112 |
| Symbol | HLA-DOB |
| Synonyms | DOB |
| Description | major histocompatibility complex, class II, DO beta |
| Reference | MIM:600629|HGNC:HGNC:4937|Ensembl:ENSG00000241106|HPRD:08368|Vega:OTTHUMG00000031213 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1.306E-5 |
| Fetal beta | -0.045 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2621419 | chr6 | 32741005 | HLA-DOB | 3112 | 0.02 | cis | ||
| rs719654 | chr6 | 32752138 | HLA-DOB | 3112 | 0.18 | cis | ||
| rs1044043 | chr6 | 32793980 | HLA-DOB | 3112 | 0.04 | cis | ||
| rs2856997 | 6 | 32781776 | HLA-DOB | ENSG00000241106.2 | 4.376E-8 | 0 | -528 | gtex_brain_putamen_basal |
| rs2856995 | 6 | 32783337 | HLA-DOB | ENSG00000241106.2 | 8.082E-9 | 0 | -2089 | gtex_brain_putamen_basal |
| rs2621326 | 6 | 32783896 | HLA-DOB | ENSG00000241106.2 | 8.082E-9 | 0 | -2648 | gtex_brain_putamen_basal |
| rs2071469 | 6 | 32784783 | HLA-DOB | ENSG00000241106.2 | 1.574E-9 | 0 | -3535 | gtex_brain_putamen_basal |
| rs2157077 | 6 | 32785904 | HLA-DOB | ENSG00000241106.2 | 1.603E-9 | 0 | -4656 | gtex_brain_putamen_basal |
| rs1894408 | 6 | 32786833 | HLA-DOB | ENSG00000241106.2 | 2.667E-8 | 0 | -5585 | gtex_brain_putamen_basal |
| rs1894407 | 6 | 32787036 | HLA-DOB | ENSG00000241106.2 | 2.669E-8 | 0 | -5788 | gtex_brain_putamen_basal |
| rs1894406 | 6 | 32787056 | HLA-DOB | ENSG00000241106.2 | 3.474E-9 | 0 | -5808 | gtex_brain_putamen_basal |
| rs4148876 | 6 | 32796793 | HLA-DOB | ENSG00000241106.2 | 9.589E-7 | 0 | -15545 | gtex_brain_putamen_basal |
| rs201194354 | 6 | 32796856 | HLA-DOB | ENSG00000241106.2 | 9.629E-7 | 0 | -15608 | gtex_brain_putamen_basal |
| rs4148875 | 6 | 32796857 | HLA-DOB | ENSG00000241106.2 | 9.622E-7 | 0 | -15609 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
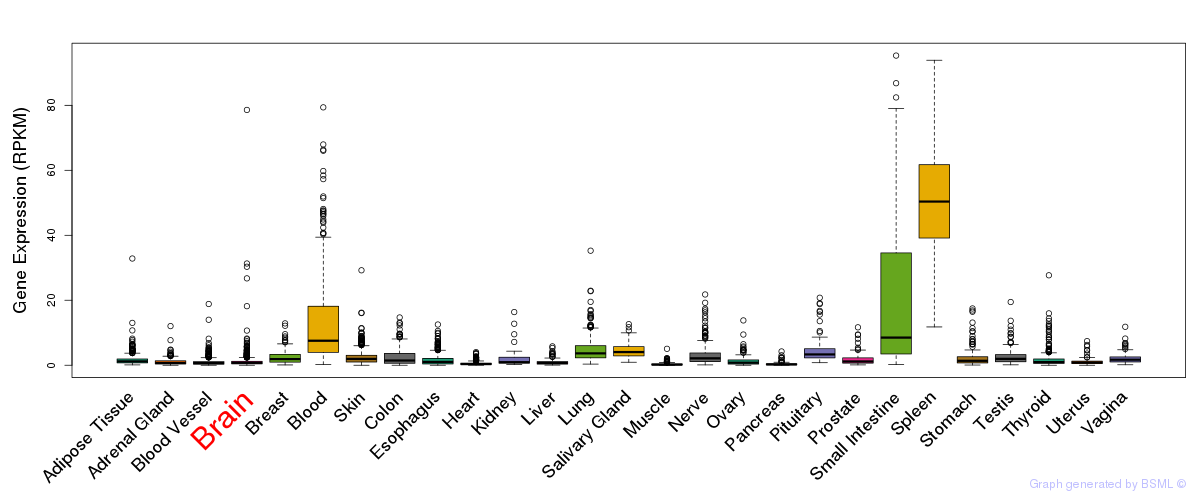
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0032395 | MHC class II receptor activity | TAS | 2998758 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005764 | lysosome | IDA | 8890155 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 11048639 |11827988 |15489916 |17652306 | |
| GO:0042613 | MHC class II protein complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG GRAFT VERSUS HOST DISEASE | 42 | 31 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC UP | 83 | 50 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS DN | 24 | 13 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4 | 61 | 34 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |