Gene Page: AOAH
Summary ?
| GeneID | 313 |
| Symbol | AOAH |
| Synonyms | - |
| Description | acyloxyacyl hydrolase |
| Reference | MIM:102593|HGNC:HGNC:548|Ensembl:ENSG00000136250|HPRD:00027|Vega:OTTHUMG00000023566 |
| Gene type | protein-coding |
| Map location | 7p14.2 |
| Pascal p-value | 0.035 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Expression | Meta-analysis of gene expression | P value: 1.788 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | AOAH | 313 | 0.18 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
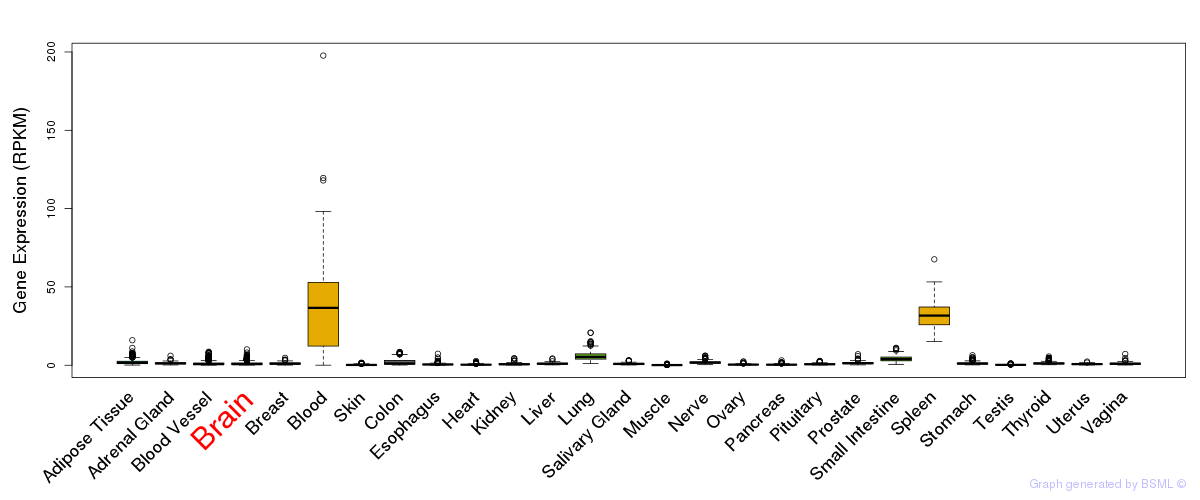
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PROS1 | 0.58 | 0.47 |
| SLC6A20 | 0.56 | 0.45 |
| DDR2 | 0.56 | 0.59 |
| LEPR | 0.55 | 0.54 |
| C7 | 0.55 | 0.49 |
| DSP | 0.54 | 0.50 |
| CYP1B1 | 0.53 | 0.54 |
| SLC47A1 | 0.52 | 0.52 |
| ITGA8 | 0.51 | 0.52 |
| OLFML1 | 0.51 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HES4 | -0.30 | -0.37 |
| AL022328.1 | -0.30 | -0.36 |
| AC120053.1 | -0.26 | -0.33 |
| RP9P | -0.26 | -0.33 |
| AF347015.21 | -0.26 | -0.21 |
| AC016757.1 | -0.25 | -0.24 |
| APOC1 | -0.24 | -0.18 |
| IL32 | -0.24 | -0.25 |
| CSAG1 | -0.24 | -0.20 |
| C16orf79 | -0.23 | -0.27 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004465 | lipoprotein lipase activity | TAS | 1883828 | |
| GO:0016788 | hydrolase activity, acting on ester bonds | IEA | - | |
| GO:0050528 | acyloxyacyl hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008653 | lipopolysaccharide metabolic process | IEA | - | |
| GO:0006629 | lipid metabolic process | TAS | 1883828 | |
| GO:0050728 | negative regulation of inflammatory response | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA UP | 33 | 28 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| MARTINELLI IMMATURE NEUTROPHIL DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE UP | 62 | 42 | All SZGR 2.0 genes in this pathway |