Gene Page: APBA1
Summary ?
| GeneID | 320 |
| Symbol | APBA1 |
| Synonyms | D9S411E|LIN10|MINT1|X11|X11A|X11ALPHA |
| Description | amyloid beta precursor protein binding family A member 1 |
| Reference | MIM:602414|HGNC:HGNC:578|Ensembl:ENSG00000107282|HPRD:03879|Vega:OTTHUMG00000019984 |
| Gene type | protein-coding |
| Map location | 9q21.12 |
| Pascal p-value | 0.004 |
| Fetal beta | -0.469 |
| Support | CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
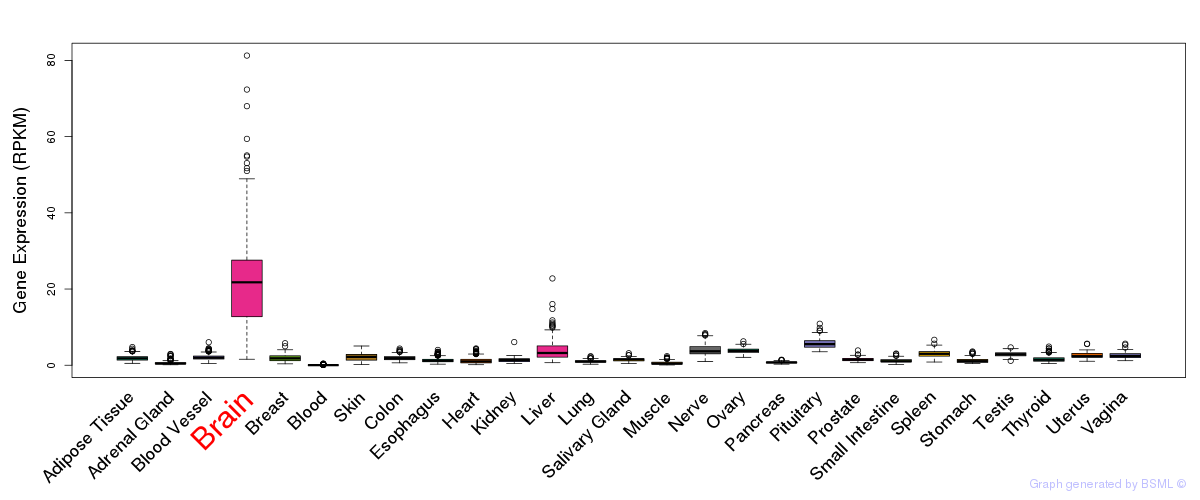
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DSTYK | 0.85 | 0.87 |
| TRAPPC10 | 0.84 | 0.86 |
| BACE1 | 0.83 | 0.86 |
| HLCS | 0.82 | 0.85 |
| DYNC1LI2 | 0.82 | 0.83 |
| CDC42BPA | 0.82 | 0.85 |
| TGOLN2 | 0.82 | 0.85 |
| GRB10 | 0.82 | 0.82 |
| C9orf5 | 0.81 | 0.84 |
| KIAA0232 | 0.81 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.59 | -0.53 |
| C1orf54 | -0.56 | -0.56 |
| C1orf61 | -0.55 | -0.59 |
| GNG11 | -0.53 | -0.50 |
| VAMP5 | -0.53 | -0.47 |
| IL32 | -0.52 | -0.49 |
| MT-CO2 | -0.51 | -0.49 |
| ENHO | -0.51 | -0.52 |
| AF347015.31 | -0.51 | -0.49 |
| C16orf74 | -0.51 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 8887653 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9753324 |
| GO:0008088 | axon cargo transport | TAS | axon (GO term level: 8) | 9822620 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9753324 |
| GO:0007155 | cell adhesion | TAS | 9753324 | |
| GO:0006461 | protein complex assembly | TAS | 9614075 |9753324 | |
| GO:0006886 | intracellular protein transport | TAS | 9614075 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | TAS | Synap, Neurotransmitter (GO term level: 12) | 9753324 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | X11alpha interacts with APP. | BIND | 11115513 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD,BioGRID | 8887653 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | APPswe interacts with X11-alpha. | BIND | 12849748 |14756819 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | Beta-APP interacts with X11. | BIND | 8887653 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | X11-ALPHA interacts with Lin-2. | BIND | 9822620 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 9753324 |9822620 |9952408|9822620 |9952408 |
| CCS | MGC138260 | copper chaperone for superoxide dismutase | - | HPRD,BioGRID | 11115513 |
| CCS | MGC138260 | copper chaperone for superoxide dismutase | X11alpha interacts with CCS. | BIND | 11115513 |
| CNTNAP3 | CASPR3 | CNTNAP3A | RP11-138L21.1 | RP11-290L7.1 | contactin associated protein-like 3 | - | HPRD,BioGRID | 12093160 |
| CNTNAP4 | CASPR4 | KIAA1763 | contactin associated protein-like 4 | - | HPRD,BioGRID | 12093160 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 14960569 |
| HTR2C | 5-HT2C | 5-HTR2C | HTR1C | 5-hydroxytryptamine (serotonin) receptor 2C | - | HPRD,BioGRID | 12006486 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 14960569 |15024025 |
| KIF17 | KIAA1405 | KIF17B | KIF3X | kinesin family member 17 | - | HPRD | 12514209 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | Affinity Capture-Western | BioGRID | 9822620 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | - | HPRD,BioGRID | 9753324 |
| LIN7C | FLJ11215 | LIN-7-C | LIN-7C | MALS-3 | MALS3 | VELI3 | lin-7 homolog C (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | Notch1 interacts with X11-alpha. This interaction was modelled on a demonstrated interaction between mouse Notch1 and human X11-alpha. | BIND | 14756819 |
| NRXN1 | DKFZp313P2036 | FLJ35941 | Hs.22998 | KIAA0578 | neurexin 1 | - | HPRD,BioGRID | 11036064 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | - | HPRD | 12196555 |
| PSEN2 | AD3L | AD4 | PS2 | STM2 | presenilin 2 (Alzheimer disease 4) | Affinity Capture-Western | BioGRID | 12196555 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD | 9395480 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | - | HPRD | 9395480 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 60 | 66 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-185 | 152 | 158 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-24 | 533 | 540 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.