Gene Page: APC
Summary ?
| GeneID | 324 |
| Symbol | APC |
| Synonyms | BTPS2|DP2|DP2.5|DP3|GS|PPP1R46 |
| Description | adenomatous polyposis coli |
| Reference | MIM:611731|HGNC:HGNC:583|Ensembl:ENSG00000134982|HPRD:01439|Vega:OTTHUMG00000128806 |
| Gene type | protein-coding |
| Map location | 5q21-q22 |
| Pascal p-value | 0.072 |
| Sherlock p-value | 0.045 |
| Fetal beta | 1.038 |
| DMG | 2 (# studies) |
| eGene | Meta |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0228 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16970232 | 5 | 112073433 | APC | -0.027 | 0.25 | DMG:Nishioka_2013 | |
| cg16481008 | 5 | 112043117 | APC | 2.29E-9 | -0.012 | 1.74E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
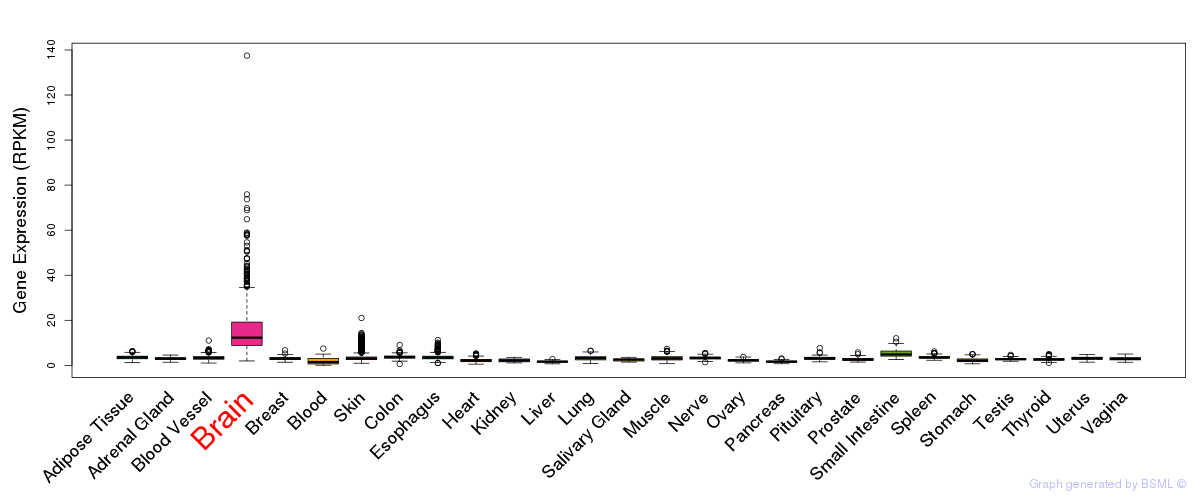
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UQCRC1 | 0.88 | 0.85 |
| NIT1 | 0.87 | 0.83 |
| ARFIP2 | 0.87 | 0.83 |
| ATP5B | 0.87 | 0.82 |
| TBRG4 | 0.86 | 0.84 |
| MRPL37 | 0.86 | 0.82 |
| NDUFA10 | 0.86 | 0.81 |
| SLC41A3 | 0.85 | 0.81 |
| PREB | 0.85 | 0.83 |
| RABGGTA | 0.85 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.62 | -0.57 |
| AF347015.21 | -0.61 | -0.50 |
| AF347015.8 | -0.61 | -0.49 |
| AL139819.3 | -0.60 | -0.59 |
| AF347015.2 | -0.59 | -0.46 |
| AF347015.26 | -0.59 | -0.49 |
| MT-CO2 | -0.58 | -0.46 |
| MT-ATP8 | -0.57 | -0.52 |
| AF347015.33 | -0.56 | -0.48 |
| MT-CYB | -0.56 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008605 | protein kinase CK2 regulator activity | IDA | 11972058 | |
| GO:0008017 | microtubule binding | IDA | 11166179 |16188939 | |
| GO:0008013 | beta-catenin binding | IPI | 11533658 | |
| GO:0008013 | beta-catenin binding | NAS | 8259518 | |
| GO:0019901 | protein kinase binding | IDA | 11283619 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | NAS | 8259518 | |
| GO:0006461 | protein complex assembly | IDA | 16188939 | |
| GO:0007026 | negative regulation of microtubule depolymerization | IDA | 11166179 | |
| GO:0009953 | dorsal/ventral pattern formation | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0007050 | cell cycle arrest | IDA | 8521819 | |
| GO:0008285 | negative regulation of cell proliferation | IDA | 8521819 | |
| GO:0009798 | axis specification | IEA | - | |
| GO:0006974 | response to DNA damage stimulus | IDA | 14728717 | |
| GO:0045736 | negative regulation of cyclin-dependent protein kinase activity | IDA | 8521819 | |
| GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | NAS | 11283619 | |
| GO:0060070 | Wnt receptor signaling pathway through beta-catenin | IC | 9601641 | |
| GO:0060070 | Wnt receptor signaling pathway through beta-catenin | NAS | 11035805 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000776 | kinetochore | IDA | 11283619 | |
| GO:0005813 | centrosome | IDA | 11283619 | |
| GO:0005829 | cytosol | EXP | 11955436 |12000790 |12820959 |15327769 |16753179 | |
| GO:0005634 | nucleus | IDA | 11035805 |12955080 | |
| GO:0005737 | cytoplasm | IDA | 11035805 |12955080 | |
| GO:0016328 | lateral plasma membrane | IDA | 12072559 | |
| GO:0030877 | beta-catenin destruction complex | IDA | 9601641 |16188939 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Affinity Capture-Western Reconstituted Complex | BioGRID | 8389242 |12070164 |
| ARHGEF4 | ASEF | ASEF1 | GEF4 | STM6 | Rho guanine nucleotide exchange factor (GEF) 4 | APC interacts with Asef. This interaction was modeled on a demonstrated interaction between human APC and Asef from an unspecified species. | BIND | 15572129 |
| ARHGEF4 | ASEF | ASEF1 | GEF4 | STM6 | Rho guanine nucleotide exchange factor (GEF) 4 | - | HPRD,BioGRID | 10947987 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Reconstituted Complex Two-hybrid | BioGRID | 9734785 |
| AXIN1 | AXIN | MGC52315 | axin 1 | - | HPRD | 11297546 |
| AXIN2 | AXIL | DKFZp781B0869 | MGC10366 | MGC126582 | axin 2 | - | HPRD,BioGRID | 10966653 |
| BUB1 | BUB1A | BUB1L | hBUB1 | budding uninhibited by benzimidazoles 1 homolog (yeast) | - | HPRD,BioGRID | 11283619 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | Biochemical Activity | BioGRID | 11283619 |
| BUB3 | BUB3L | hBUB3 | budding uninhibited by benzimidazoles 3 homolog (yeast) | Affinity Capture-Western | BioGRID | 11283619 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | - | HPRD | 9637688 |9811605 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 11972058 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | Affinity Capture-Western | BioGRID | 11972058 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | CtBP interacts with APC. | BIND | 15525529 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | Affinity Capture-Western | BioGRID | 7651399 |8259519 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | APC interacts with Beta-catenin. | BIND | 15525529 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 12000790 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Beta-catenin interacts with APC. This interaction was modelled on a demonstrated interaction between human beta-catenin and mouse Apc. | BIND | 15294866 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western Co-crystal Structure Far Western Reconstituted Complex | BioGRID | 8259519 |9286858 |11251183 |11533658 |11707392 |11712088 |11972058 |12628243 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9188857 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD,BioGRID | 9286858 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-Western | BioGRID | 11251183 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | APC interacts with IQGAP1. | BIND | 15572129 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Affinity Capture-Western Reconstituted Complex | BioGRID | 7651399 |8074697 |
| KIFAP3 | FLJ22818 | KAP3 | SMAP | Smg-GDS | dJ190I16.1 | kinesin-associated protein 3 | - | HPRD,BioGRID | 11912492 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD,BioGRID | 9637688 |
| MAPRE1 | EB1 | MGC117374 | MGC129946 | microtubule-associated protein, RP/EB family, member 1 | - | HPRD,BioGRID | 11470413 |12388762 |
| MAPRE2 | EB1 | EB2 | RP1 | microtubule-associated protein, RP/EB family, member 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7606712 |11470413 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | Affinity Capture-Western | BioGRID | 15020226 |
| PPP2R5A | B56A | MGC131915 | PR61A | protein phosphatase 2, regulatory subunit B', alpha isoform | Co-localization | BioGRID | 11585828 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 7744883 |
| PTPN13 | DKFZp686J1497 | FAP-1 | PNP1 | PTP-BAS | PTP-BL | PTP1E | PTPL1 | PTPLE | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | Affinity Capture-Western Two-hybrid | BioGRID | 10951583 |
| RP1 | DCDC4A | ORP1 | retinitis pigmentosa 1 (autosomal dominant) | - | HPRD | 10188731 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | - | HPRD,BioGRID | 11389840 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Affinity Capture-Western Reconstituted Complex | BioGRID | 15331612 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | Co-localization Reconstituted Complex | BioGRID | 11166179 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12070164 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PS1 PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN DEG PATHWAY | 18 | 17 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CTNNB1 PHOSPHORYLATION CASCADE | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING DN | 58 | 35 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 27 | 15 | 8 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE DN | 33 | 22 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS DIRECT DN | 14 | 7 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL SHORT TERM | 32 | 15 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C3 | 14 | 10 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C CLUSTER UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA | 43 | 27 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 DN | 69 | 38 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 382 | 388 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-129-5p | 404 | 411 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-142-3p | 310 | 317 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-153 | 961 | 967 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| hsa-miR-153 | UUGCAUAGUCACAAAAGUGA | ||||
| miR-155 | 965 | 971 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-203.1 | 822 | 829 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-26 | 1248 | 1255 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 462 | 468 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-3p | 859 | 865 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-330 | 511 | 517 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-374 | 359 | 365 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 741 | 747 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-448 | 401 | 407 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-450 | 404 | 410 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-496 | 847 | 853 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.