Gene Page: HSD11B1
Summary ?
| GeneID | 3290 |
| Symbol | HSD11B1 |
| Synonyms | 11-DH|11-beta-HSD1|CORTRD2|HDL|HSD11|HSD11B|HSD11L|SDR26C1 |
| Description | hydroxysteroid (11-beta) dehydrogenase 1 |
| Reference | MIM:600713|HGNC:HGNC:5208|Ensembl:ENSG00000117594|HPRD:07042|Vega:OTTHUMG00000036481 |
| Gene type | protein-coding |
| Map location | 1q32-q41 |
| Fetal beta | -1.333 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.851 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4120839 | chr1 | 181816818 | HSD11B1 | 3290 | 0.02 | trans | ||
| rs2275869 | chr1 | 205274605 | HSD11B1 | 3290 | 0.04 | trans | ||
| rs17572651 | chr1 | 218943612 | HSD11B1 | 3290 | 0.02 | trans | ||
| snp_a-1919794 | 0 | HSD11B1 | 3290 | 0.08 | trans | |||
| rs12622326 | chr2 | 7894385 | HSD11B1 | 3290 | 0.19 | trans | ||
| rs16829545 | chr2 | 151977407 | HSD11B1 | 3290 | 5.021E-7 | trans | ||
| rs7584986 | chr2 | 184111432 | HSD11B1 | 3290 | 6.216E-6 | trans | ||
| rs17008051 | chr3 | 70998485 | HSD11B1 | 3290 | 0 | trans | ||
| rs13439821 | chr8 | 116213885 | HSD11B1 | 3290 | 0.07 | trans | ||
| rs16887186 | chr8 | 116231703 | HSD11B1 | 3290 | 0.05 | trans | ||
| rs990438 | chr15 | 24322236 | HSD11B1 | 3290 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | HSD11B1 | 3290 | 2.235E-9 | trans |
Section II. Transcriptome annotation
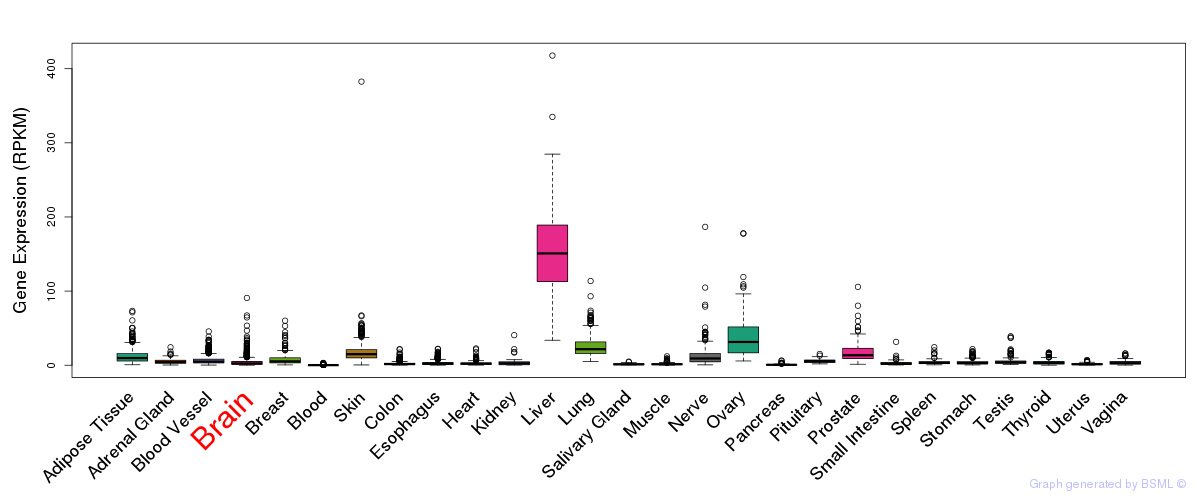
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAJB1 | 0.95 | 0.57 |
| HSPA1A | 0.94 | 0.56 |
| HSPA1B | 0.91 | 0.62 |
| G0S2 | 0.82 | 0.29 |
| BAG3 | 0.82 | 0.26 |
| AL031847.1 | 0.71 | 0.12 |
| ACRC | 0.66 | 0.07 |
| DNAJB4 | 0.59 | 0.04 |
| HSPB1 | 0.57 | 0.25 |
| C1orf51 | 0.56 | 0.21 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FOXS1 | -0.24 | 0.10 |
| SLC5A6 | -0.23 | 0.16 |
| WDR81 | -0.23 | 0.10 |
| ARHGEF15 | -0.22 | 0.07 |
| CD34 | -0.22 | -0.03 |
| ACVRL1 | -0.22 | 0.08 |
| DNM2 | -0.22 | 0.17 |
| IFNAR2 | -0.21 | 0.18 |
| MYLK2 | -0.21 | -0.00 |
| SLC38A5 | -0.21 | 0.11 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003845 | 11-beta-hydroxysteroid dehydrogenase activity | EXP | 1885595 | |
| GO:0003845 | 11-beta-hydroxysteroid dehydrogenase activity | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006694 | steroid biosynthetic process | EXP | 15583024 | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0030324 | lung development | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | EXP | 1885595 | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STEROID HORMONE BIOSYNTHESIS | 55 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| PIEPOLI LGI1 TARGETS DN | 14 | 6 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| MOROSETTI FACIOSCAPULOHUMERAL MUSCULAR DISTROPHY UP | 20 | 11 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 DN | 65 | 39 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| WANG IMMORTALIZED BY HOXA9 AND MEIS1 UP | 31 | 24 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| WANG CLASSIC ADIPOGENIC TARGETS OF PPARG | 26 | 15 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY DN | 61 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-326 | 98 | 105 | 1A,m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.