Gene Page: HSPA5
Summary ?
| GeneID | 3309 |
| Symbol | HSPA5 |
| Synonyms | BIP|GRP78|HEL-S-89n|MIF2 |
| Description | heat shock protein family A (Hsp70) member 5 |
| Reference | MIM:138120|HGNC:HGNC:5238|Ensembl:ENSG00000044574|HPRD:00682|Vega:OTTHUMG00000020672 |
| Gene type | protein-coding |
| Map location | 9q33.3 |
| Pascal p-value | 0.549 |
| Sherlock p-value | 0.053 |
| Fetal beta | 0.241 |
| eGene | Myers' cis & trans |
| Support | RNA AND PROTEIN SYNTHESIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.7656 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10868543 | chr9 | 89852686 | HSPA5 | 3309 | 0.16 | trans | ||
| rs7331934 | chr13 | 29698636 | HSPA5 | 3309 | 0.15 | trans |
Section II. Transcriptome annotation
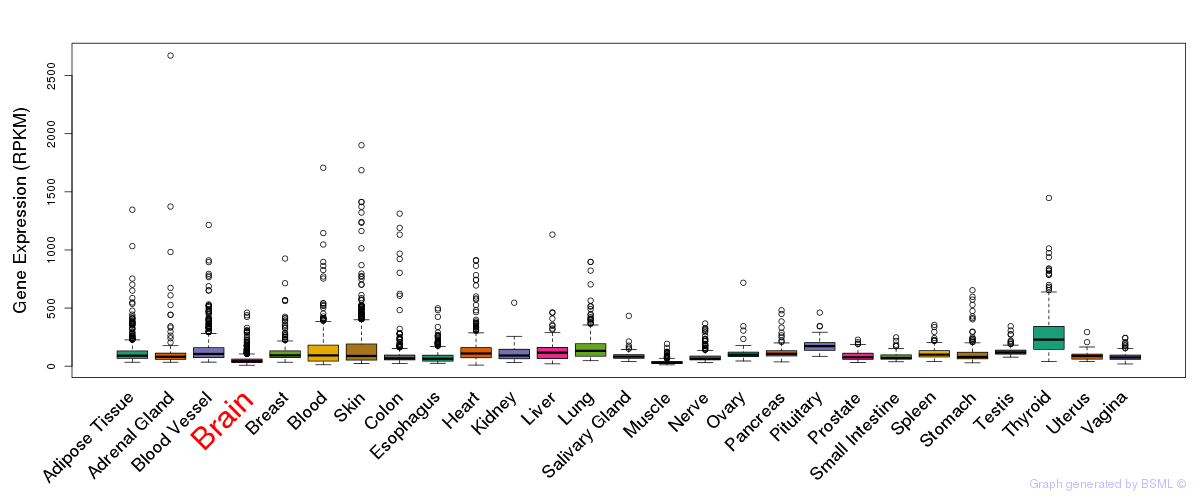
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| XRCC6 | 0.89 | 0.89 |
| DDX50 | 0.89 | 0.90 |
| DNAJA1 | 0.88 | 0.88 |
| CPSF3 | 0.88 | 0.90 |
| CCT5 | 0.88 | 0.90 |
| ME2 | 0.88 | 0.88 |
| ST13 | 0.88 | 0.87 |
| TARS | 0.88 | 0.89 |
| C22orf28 | 0.87 | 0.86 |
| CCT6A | 0.87 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.68 | -0.76 |
| AF347015.31 | -0.68 | -0.76 |
| IFI27 | -0.68 | -0.76 |
| AF347015.33 | -0.67 | -0.77 |
| AF347015.21 | -0.67 | -0.74 |
| AF347015.27 | -0.66 | -0.74 |
| AF347015.8 | -0.66 | -0.75 |
| FXYD1 | -0.65 | -0.74 |
| HIGD1B | -0.65 | -0.74 |
| MT-CYB | -0.64 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 16130169 | |
| GO:0005524 | ATP binding | NAS | 12665508 | |
| GO:0030674 | protein binding, bridging | NAS | - | |
| GO:0051082 | unfolded protein binding | TAS | 16130169 | |
| GO:0043022 | ribosome binding | IEA | - | |
| GO:0043027 | caspase inhibitor activity | IDA | 12665508 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006983 | ER overload response | IEA | - | |
| GO:0006916 | anti-apoptosis | IMP | 12665508 | |
| GO:0006916 | anti-apoptosis | TAS | 16130169 | |
| GO:0042149 | cellular response to glucose starvation | IDA | 10085239 | |
| GO:0043154 | negative regulation of caspase activity | IDA | 12665508 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005793 | ER-Golgi intermediate compartment | IDA | 15308636 | |
| GO:0005788 | endoplasmic reticulum lumen | TAS | 12665508 | |
| GO:0005634 | nucleus | IDA | 12665508 | |
| GO:0005783 | endoplasmic reticulum | TAS | 16130169 | |
| GO:0009986 | cell surface | IDA | 12493773 | |
| GO:0008303 | caspase complex | IDA | 12665508 | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 12665508 | |
| GO:0030176 | integral to endoplasmic reticulum membrane | IDA | 12665508 | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A2M | CPAMD5 | DKFZp779B086 | FWP007 | S863-7 | alpha-2-macroglobulin | - | HPRD,BioGRID | 12194978 |
| APOB | FLDB | apolipoprotein B (including Ag(x) antigen) | - | HPRD | 12397072 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Two-hybrid | BioGRID | 16169070 |
| CASP12 | CASP12P1 | caspase 12 (gene/pseudogene) | Affinity Capture-Western | BioGRID | 11943137 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11943137 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| DNAJC1 | DNAJL1 | ERdj1 | HTJ1 | MGC131954 | MTJ1 | DnaJ (Hsp40) homolog, subfamily C, member 1 | - | HPRD | 10777498 |
| DNAJC10 | DKFZp434J1813 | ERdj5 | JPDI | MGC104194 | DnaJ (Hsp40) homolog, subfamily C, member 10 | - | HPRD,BioGRID | 12411443 |
| DPH1 | DPH2L | DPH2L1 | FLJ33211 | OVCA1 | DPH1 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11013075 |
| EIF2AK3 | DKFZp781H1925 | HRI | PEK | PERK | WRS | eukaryotic translation initiation factor 2-alpha kinase 3 | - | HPRD,BioGRID | 11907036 |
| ERN1 | FLJ30999 | IRE1 | IRE1P | MGC163277 | MGC163279 | endoplasmic reticulum to nucleus signaling 1 | - | HPRD,BioGRID | 12637535 |
| ERP29 | C12orf8 | ERp28 | ERp31 | PDI-DB | endoplasmic reticulum protein 29 | - | HPRD | 9492298 |
| F8 | AHF | DXS1253E | F8B | F8C | FVIII | HEMA | coagulation factor VIII, procoagulant component | - | HPRD | 9305856 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD | 10461883 |
| HLA-C | D6S204 | FLJ27082 | HLA-Cw | HLA-Cw12 | HLA-JY3 | HLC-C | PSORS1 | major histocompatibility complex, class I, C | - | HPRD,BioGRID | 7957306 |
| HTR3A | 5-HT-3 | 5-HT3A | 5-HT3R | 5HT3R | HTR3 | 5-hydroxytryptamine (serotonin) receptor 3A | - | HPRD | 12359150 |
| IGHM | DKFZp686I15196 | DKFZp686I15212 | FLJ00385 | MGC104996 | MGC52291 | MU | VH | immunoglobulin heavy constant mu | - | HPRD | 3122216 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 9409741 |
| KRT8 | CARD2 | CK8 | CYK8 | K2C8 | K8 | KO | keratin 8 | - | HPRD,BioGRID | 9409741 |
| LCT | LAC | LPH | LPH1 | lactase | - | HPRD,BioGRID | 11751874 |
| LDLR | FH | FHC | low density lipoprotein receptor | - | HPRD,BioGRID | 10906332 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 14743216 |
| PCSK7 | LPC | PC7 | PC8 | SPC7 | proprotein convertase subtilisin/kexin type 7 | - | HPRD,BioGRID | 10964928 |
| PRNP | ASCR | CD230 | CJD | GSS | MGC26679 | PRIP | PrP | PrP27-30 | PrP33-35C | PrPc | prion | prion protein | PrPc interacts with HSPA5 (BiP). | BIND | 10970892 |
| PSME3 | Ki | PA28-gamma | PA28G | REG-GAMMA | proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) | Two-hybrid | BioGRID | 16169070 |
| SIL1 | BAP | MSS | ULG5 | SIL1 homolog, endoplasmic reticulum chaperone (S. cerevisiae) | - | HPRD,BioGRID | 12356756 |
| STMN1 | LAP18 | Lag | OP18 | PP17 | PP19 | PR22 | SMN | stathmin 1/oncoprotein 18 | - | HPRD,BioGRID | 7724523 |10197448 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
| TG | AITD3 | TGN | thyroglobulin | - | HPRD,BioGRID | 10049727 |11294872 |
| TMEM132A | DKFZp547E212 | FLJ20539 | GBP | HSPA5BP1 | MGC138669 | transmembrane protein 132A | - | HPRD,BioGRID | 12514190 |
| TRA@ | FLJ22602 | MGC117436 | MGC22624 | MGC23964 | MGC71411 | TCRA | TCRD | TRA | T cell receptor alpha locus | - | HPRD | 1649196 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | TSHR interacts with BiP. This interaction was modeled on a demonstrated interaction between human TSHR and hamster BiP. | BIND | 12383251 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD,BioGRID | 12383251 |
| VWF | F8VWF | VWD | von Willebrand factor | - | HPRD,BioGRID | 10887119 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PROTEIN EXPORT | 24 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| NEBEN AML WITH FLT3 OR NRAS DN | 12 | 6 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME DN | 17 | 9 | All SZGR 2.0 genes in this pathway |
| GOTTWEIN TARGETS OF KSHV MIR K12 11 | 63 | 45 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| HONMA DOCETAXEL RESISTANCE | 34 | 23 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS DN | 17 | 14 | All SZGR 2.0 genes in this pathway |
| NGO MALIGNANT GLIOMA 1P LOH | 17 | 13 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-181 | 84 | 90 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-199 | 345 | 351 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-30-5p | 142 | 148 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-378 | 41 | 47 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-379 | 309 | 315 | 1A | hsa-miR-379brain | UGGUAGACUAUGGAACGUA |
| miR-495 | 334 | 341 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.