Gene Page: IL9
Summary ?
| GeneID | 3578 |
| Symbol | IL9 |
| Synonyms | HP40|IL-9|P40 |
| Description | interleukin 9 |
| Reference | MIM:146931|HGNC:HGNC:6029|Ensembl:ENSG00000145839|HPRD:00910|Vega:OTTHUMG00000129147 |
| Gene type | protein-coding |
| Map location | 5q31.1 |
| Pascal p-value | 0.141 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
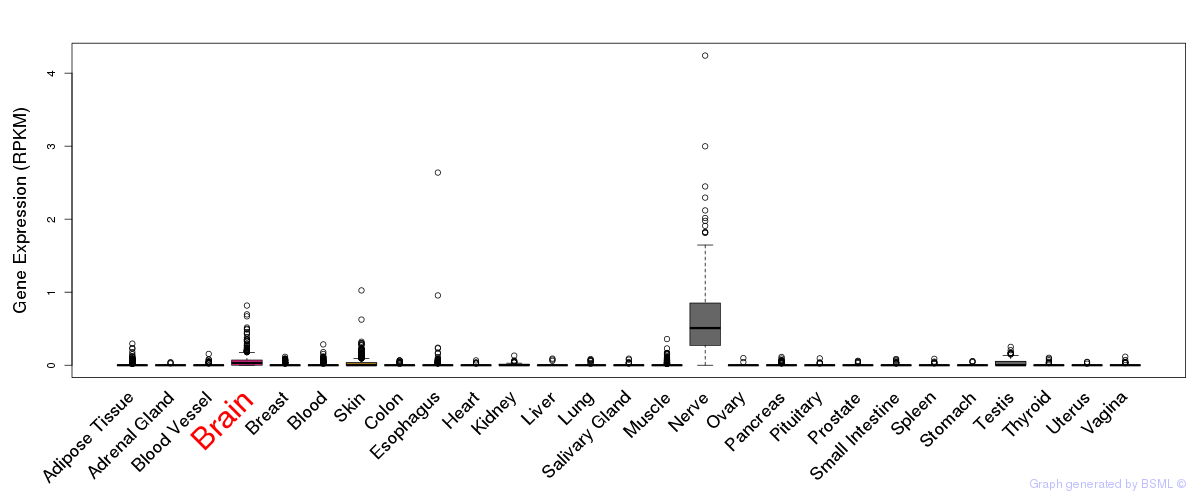
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HYDIN | 0.59 | 0.53 |
| C9orf117 | 0.58 | 0.41 |
| RP1-199H16.1 | 0.57 | 0.45 |
| DNAH12 | 0.56 | 0.39 |
| AGBL2 | 0.56 | 0.29 |
| VWA3A | 0.56 | 0.15 |
| SPEF2 | 0.56 | 0.44 |
| C3orf15 | 0.55 | 0.50 |
| AC002472.1 | 0.55 | 0.22 |
| CCDC37 | 0.55 | 0.47 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.38 | -0.45 |
| AF347015.21 | -0.36 | -0.46 |
| AF347015.31 | -0.36 | -0.45 |
| AF347015.27 | -0.36 | -0.42 |
| AF347015.8 | -0.36 | -0.43 |
| AF347015.2 | -0.35 | -0.41 |
| MT-CYB | -0.34 | -0.41 |
| AF347015.33 | -0.33 | -0.39 |
| PLA2G5 | -0.33 | -0.43 |
| IL32 | -0.33 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005126 | hematopoietin/interferon-class (D200-domain) cytokine receptor binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008284 | positive regulation of cell proliferation | TAS | 1680606 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006954 | inflammatory response | TAS | 9371819 | |
| GO:0045407 | positive regulation of interleukin-5 biosynthetic process | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERYTH PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STEM PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 2 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES T1 | 9 | 7 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |