Gene Page: ARL4D
Summary ?
| GeneID | 379 |
| Symbol | ARL4D |
| Synonyms | ARF4L|ARL6 |
| Description | ADP ribosylation factor like GTPase 4D |
| Reference | MIM:600732|HGNC:HGNC:656|Ensembl:ENSG00000175906|HPRD:02842|Vega:OTTHUMG00000180881 |
| Gene type | protein-coding |
| Map location | 17q21.31 |
| Pascal p-value | 0.081 |
| Sherlock p-value | 8.738E-4 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.18:CC_BA10_disease_P=0.0028:HBB_BA9_fold_change=-1.20:HBB_BA9_disease_P=0.0469 |
| Fetal beta | 0.122 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19252218 | 17 | 41477271 | ARL4D | 3.901E-4 | 0.353 | 0.043 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16827037 | chr3 | 117203284 | ARL4D | 379 | 0.04 | trans | ||
| rs7029518 | chr9 | 37782560 | ARL4D | 379 | 0.12 | trans | ||
| rs17761129 | chr15 | 24262387 | ARL4D | 379 | 0.15 | trans | ||
| rs9319807 | chr18 | 68766779 | ARL4D | 379 | 0.02 | trans | ||
| rs16978981 | chrX | 13538144 | ARL4D | 379 | 0.07 | trans |
Section II. Transcriptome annotation
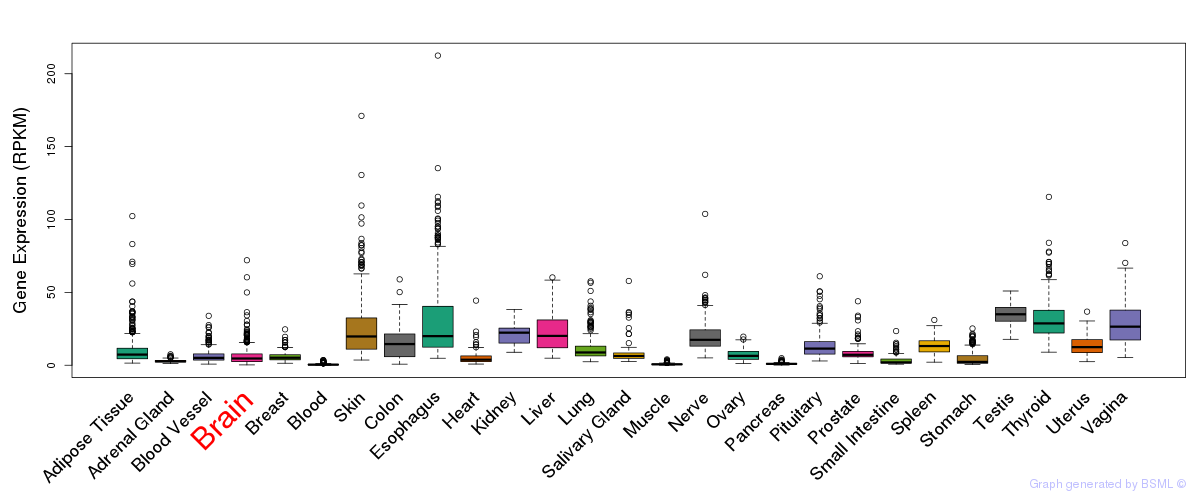
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1609 | 0.94 | 0.92 |
| C20orf11 | 0.94 | 0.92 |
| GTPBP1 | 0.94 | 0.92 |
| TAF5L | 0.94 | 0.92 |
| CHMP7 | 0.93 | 0.94 |
| NUP62 | 0.93 | 0.94 |
| ZNF71 | 0.93 | 0.93 |
| ZNF746 | 0.93 | 0.94 |
| POLDIP3 | 0.93 | 0.92 |
| GEMIN4 | 0.93 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.75 | -0.89 |
| AF347015.31 | -0.75 | -0.87 |
| AF347015.27 | -0.75 | -0.87 |
| C5orf53 | -0.75 | -0.78 |
| AF347015.33 | -0.74 | -0.87 |
| FXYD1 | -0.74 | -0.85 |
| S100B | -0.74 | -0.84 |
| AF347015.8 | -0.72 | -0.87 |
| MT-CYB | -0.72 | -0.85 |
| AC021016.1 | -0.70 | -0.83 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARL6IP1 | AIP1 | ARL6IP | ARMER | KIAA0069 | ADP-ribosylation factor-like 6 interacting protein 1 | - | HPRD | 10508919 |
| DNAJA1 | DJ-2 | DjA1 | HDJ2 | HSDJ | HSJ2 | HSPF4 | hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 | Two-hybrid | BioGRID | 16169070 |
| EIF2B1 | EIF-2B | EIF-2Balpha | EIF2B | EIF2BA | MGC117409 | MGC125868 | MGC125869 | eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa | Two-hybrid | BioGRID | 16169070 |
| EML4 | C2orf2 | DKFZp686P18118 | ELP120 | FLJ10942 | FLJ32318 | ROPP120 | echinoderm microtubule associated protein like 4 | Two-hybrid | BioGRID | 16169070 |
| EPRS | DKFZp313B047 | EARS | GLUPRORS | PARS | PIG32 | QARS | QPRS | glutamyl-prolyl-tRNA synthetase | Two-hybrid | BioGRID | 16169070 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | Two-hybrid | BioGRID | 16169070 |
| NDRG1 | CAP43 | CMT4D | DRG1 | GC4 | HMSNL | NDR1 | NMSL | PROXY1 | RIT42 | RTP | TARG1 | TDD5 | N-myc downstream regulated 1 | Two-hybrid | BioGRID | 16169070 |
| PGAM1 | PGAM-B | PGAMA | phosphoglycerate mutase 1 (brain) | Two-hybrid | BioGRID | 16169070 |
| PRKCSH | AGE-R2 | G19P1 | PCLD | PLD1 | protein kinase C substrate 80K-H | Two-hybrid | BioGRID | 16169070 |
| SNRPN | DKFZp686C0927 | DKFZp686M12165 | DKFZp761I1912 | DKFZp762N022 | FLJ33569 | FLJ36996 | FLJ39265 | HCERN3 | MGC29886 | PWCR | RT-LI | SM-D | SMN | SNRNP-N | SNURF-SNRPN | small nuclear ribonucleoprotein polypeptide N | Two-hybrid | BioGRID | 16169070 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| UBR1 | JBS | MGC142065 | MGC142067 | ubiquitin protein ligase E3 component n-recognin 1 | Two-hybrid | BioGRID | 16169070 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI RESPONSE TO ROMIDEPSIN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 8HR | 39 | 31 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION | 37 | 20 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |